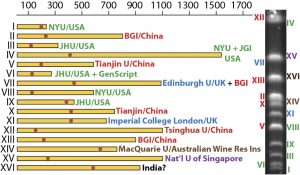
Following our announcement this month of a new collaboration and integration with protocols.io, we’ve gone into more detail on the first two papers that have utilised this open access repository of scientific methods and collaborative protocol-centered platform.