
FUNC on Safari: Functional Metagenomics conference 2024 at Skukuza, Kruger National Park, South Africa (2nd – 5th June 2024) FMG24 participants group photo, taken outside the conference centre at Skukuza rest camp.

FUNC on Safari: Functional Metagenomics conference 2024 at Skukuza, Kruger National Park, South Africa (2nd – 5th June 2024) FMG24 participants group photo, taken outside the conference centre at Skukuza rest camp.

This week in GigaScience we published research revealing a previously hidden diversity of symbiotic bacteria from the genus Rickettsia , spanning a wide range of arthropod hosts. Using accidentally amplified sequence data from a barcoding database as a starting point, the results will help to better understand the co-evolution of these intimate symbioses.

Yes, this is a clickbait headline, and yes, it may seem like shooting fish in a barrel to complain about crappy data in GBIF, but my point here is raise concerns about the impact of metagenomic data on GBIF, and how difficult it may be to track down the causes of errors.

In this data-driven era, research is faced with new challenges, from sharing, storing and accessing data, including how to better integrate data to answer big questions in science. With many data repositories available, it is hard to maintain them all – some repositories are forced to close – meaning loss of access to invaluable datasets.

Photograph by Giang-Son Nguyen (SINTEF) The Norwegian city of Trondheim seemed the place to be this June – with hotels in high demand

The mock metagenome, MAGs and breaking the first rule of Long Read Club Nick showing us some of his experiments with an early antecedent Short Read Club… Out today in GigaScience is a new “mock metagenome” Data Note from the Nick Loman lab in Birmingham showcasing the latest long-read sequencing technologies from Oxford Nanopore.

Out today is the winner of our ICG13 Prize, presenting work that can aid in revealing new biologically relevant findings and missed genes from previously generated transcriptome assemblies. Teaching old data new tricks, and maximising every last nugget of information from previously funded research.
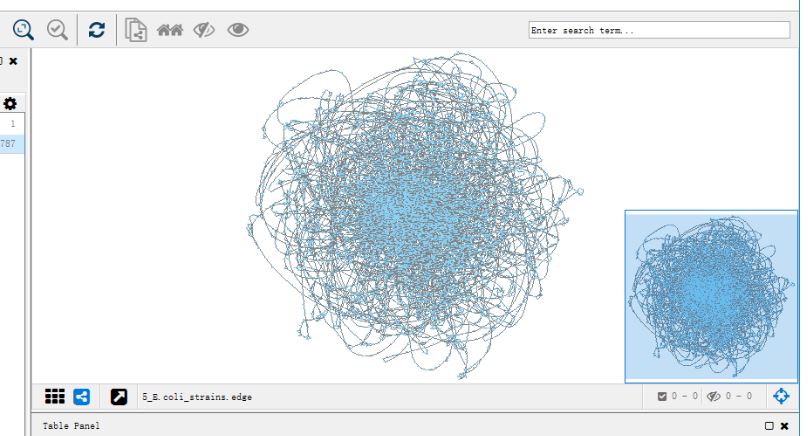
Navigating Pangenome’s Labyrinth In the two decades since the first genomes were sequenced, with the exponential growth of new and closely related genomes it has become increasingly difficult to visualise and compare their structure. Particularly with the large diversity and difference in genes within microbial genomes.
Monkey Microbiome Business Research just out in GigaScience introduces the macaque monkey to the microbial gene catalogue club, joining other important model organisms including the (also recently published in GigaScience) Rat, Mouse, Pig and Cow.
** GigaScience has Tapeworms and Scabies! And Reproducible Research. **While there has been recent controversy (and hashtags in response) from some of the more conservative sections of the medical community calling those who use or build on previous data “research parasites”, as data publishers we strongly disagree with this.