At rOpenSci, much of our code, content and infrastructure is hosted on GitHub over several organizations – described on our resources page.This post summarizes some steps we’ve taken to safeguard our GitHub organizations.Paying attention to access rights &
Publicaciones de Rogue Scholar
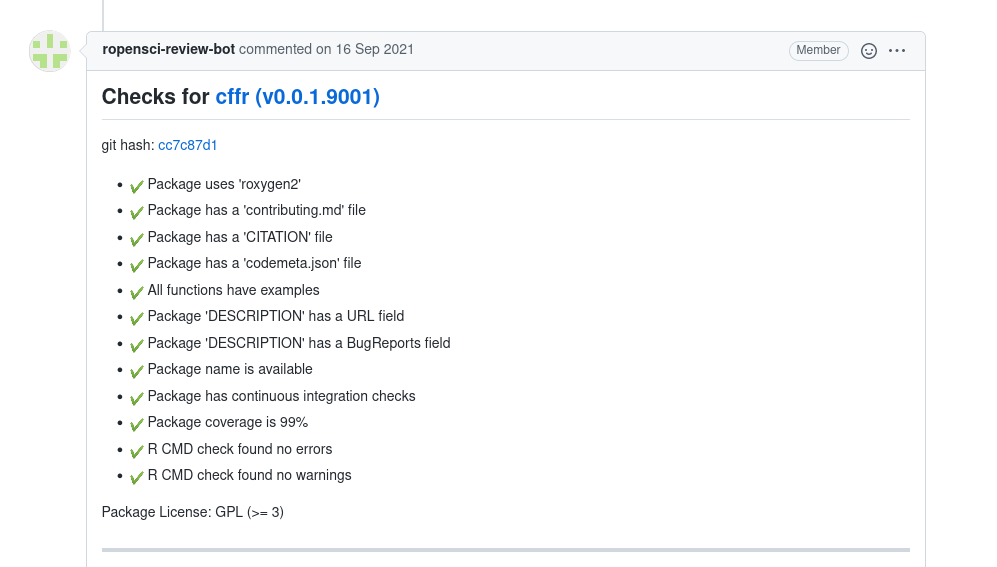
What is pkgcheck? The pkgcheck package hasbeen developed by rOpenSci to automate the process of checking all packages onsubmission.
The rOpenSci R-universe is a bit special as, compared to other R-universes, it builds docs for all the packages in our suite.Looking at the dashboard helps us identify failures in building the packages as well as in building the pkgdown websites.We then help authors fix these issues in order to comply with our package curation policy.As a package author you should also rely on continuous integration in your own repo for catching e.g. R CMD check
What is renv RStudio’s renv package is a powerful dependency management toolkit for R. It allows you to create a lockfile that records the exact versions of R packages used in a given project, and provides tooling to install exactly those same versions on another machine, or at a later point in time. This is very useful to create an isolated project environment for reproducibility or production purposes.
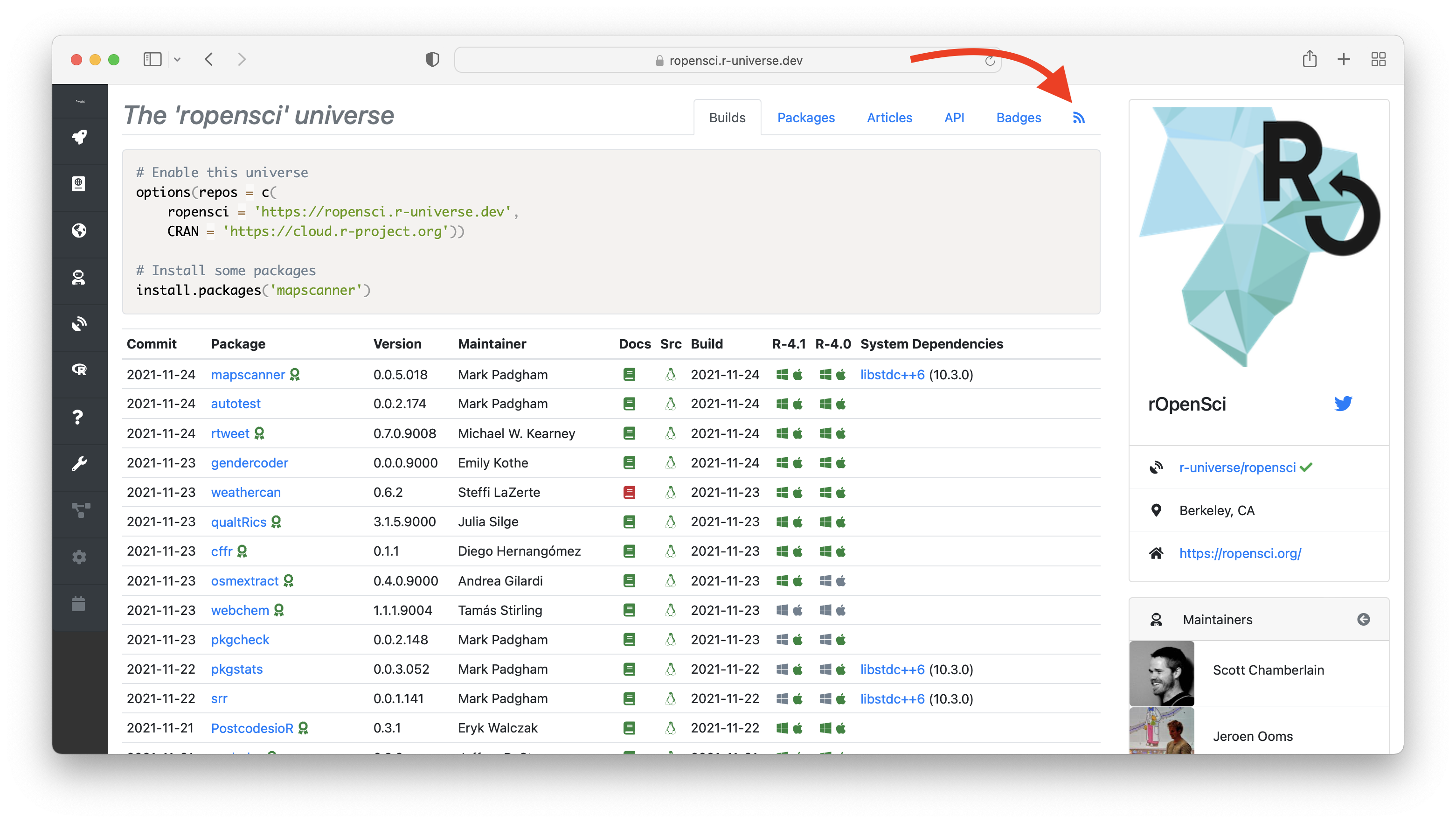
Continuous deployment in r-universe A major difference between r-universe and static repositories like CRAN and BioConductor is continuous deployment: packages in r-universe are continuously built in CI and immediately deployed to our package server. This package server stores binaries and metadata in a database, which enables us to dynamically query and expose all the package data through APIs, dashboards, feeds, etc.
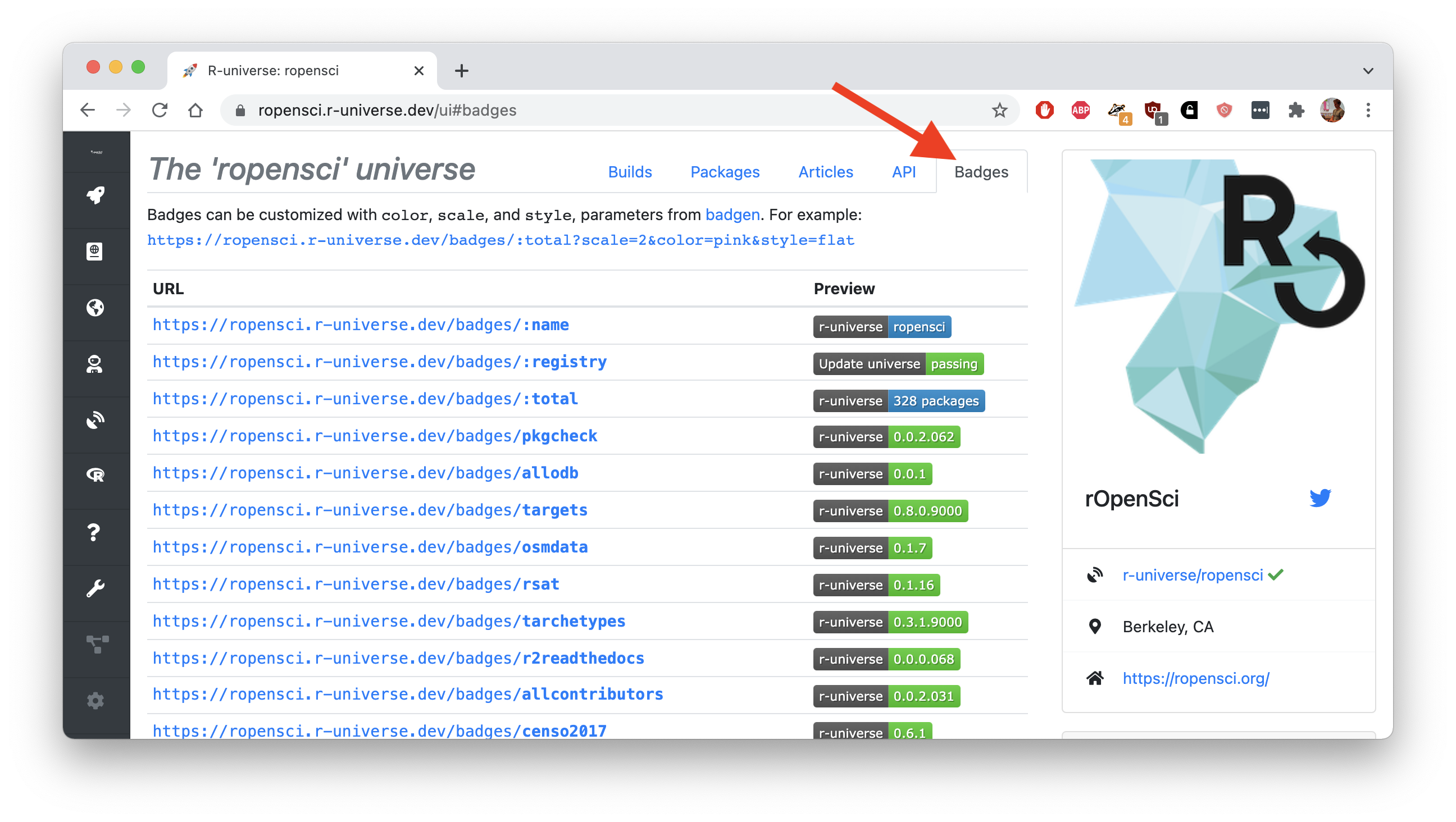
Badges in r-universe After creating your personal universe, the dashboard on https://{yourname}.r-universe.dev shows the version and other details for each package in your repository. We have also added a new tab that lists the available badges for the repository.
Have you ever found a command-line tool that’s perfect for getting your job done, and wanted to use it from an R script or package?E.g. some sort of scientific software providing a specific functionality made available though a command-line interface (CLI)? In this post, we have a look at a few options for writing such CLI wrappers in R.In particular, we compare the base R functions system() and system2(), the sys package and the processx
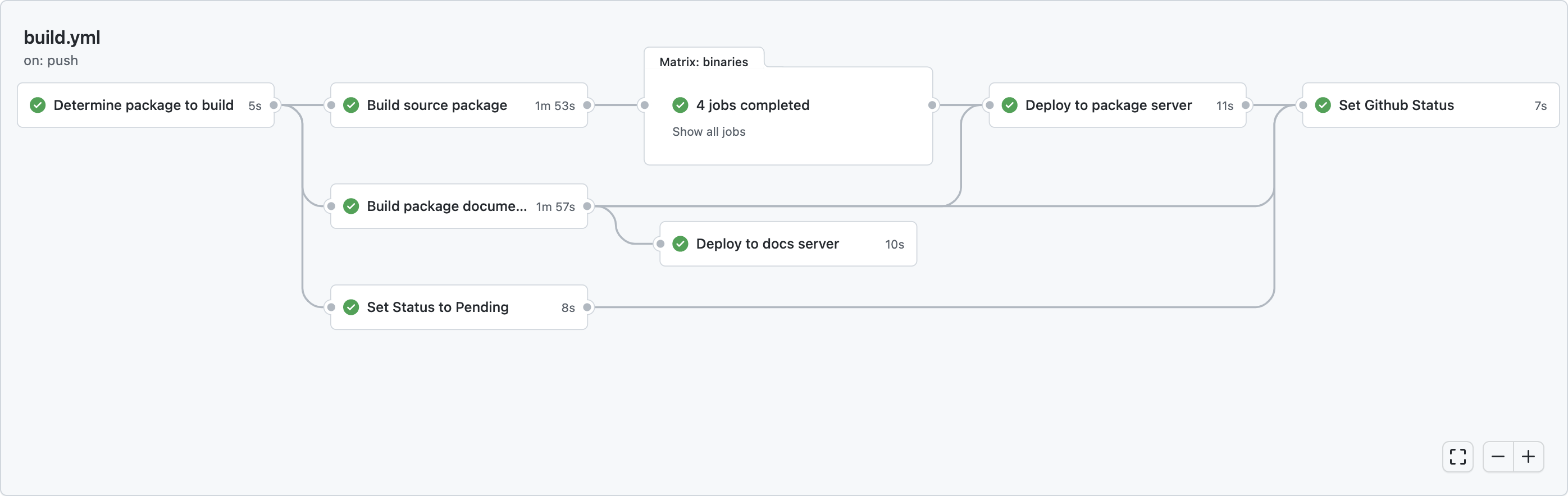
TLDR: As of today, the pkgdown sites for all rOpenSci packages on our docs server are built on r-universe. This is an internal change; no action is required for package maintainers.From Jenkins to r-universe For the past 2 years we have been using a Jenkins server to automatically build the pkgdown sites for all rOpenSci packages, which get published on https://docs.ropensci.org.
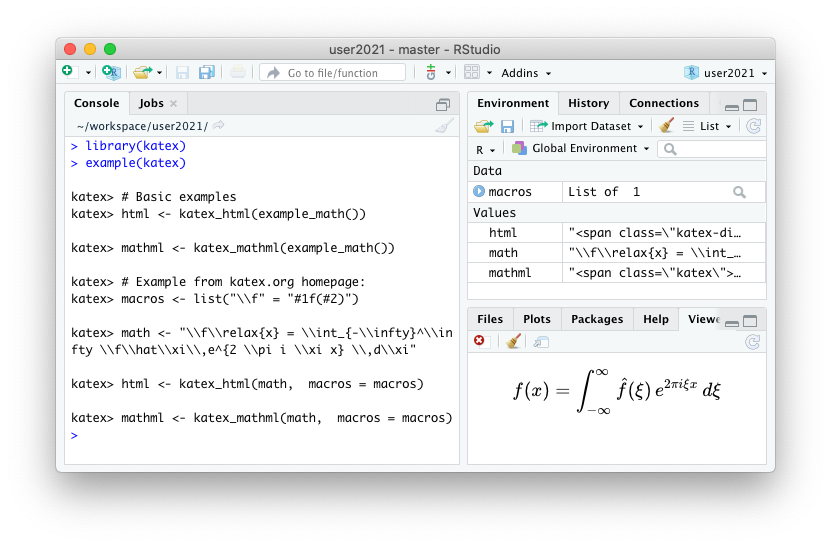
A new rOpenSci package katex is now on CRAN.This package allows for converting latex math expressions to HTML and MathML for use in markdown documents or package documentation.
How to keep up with rOpenSci?We agree that we’re doing so much good work that it’s hard.