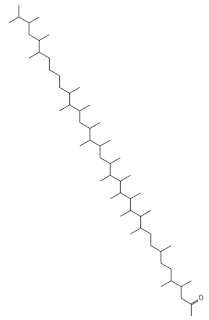
About two weeks ago, the ChemConnector blog reported an InChIKey collosion detected by Prof. Goodman. Unlike the previous collision, this one was based solely on the graph and not on stereochemistry. The two molecules both have the InChIKey OCPAUTFLLNMYSX-UHFFFAOYSA-N: The compounds are really different, the molecular formulas are C 50 H 102 O and C 57 H 114 O respectively.