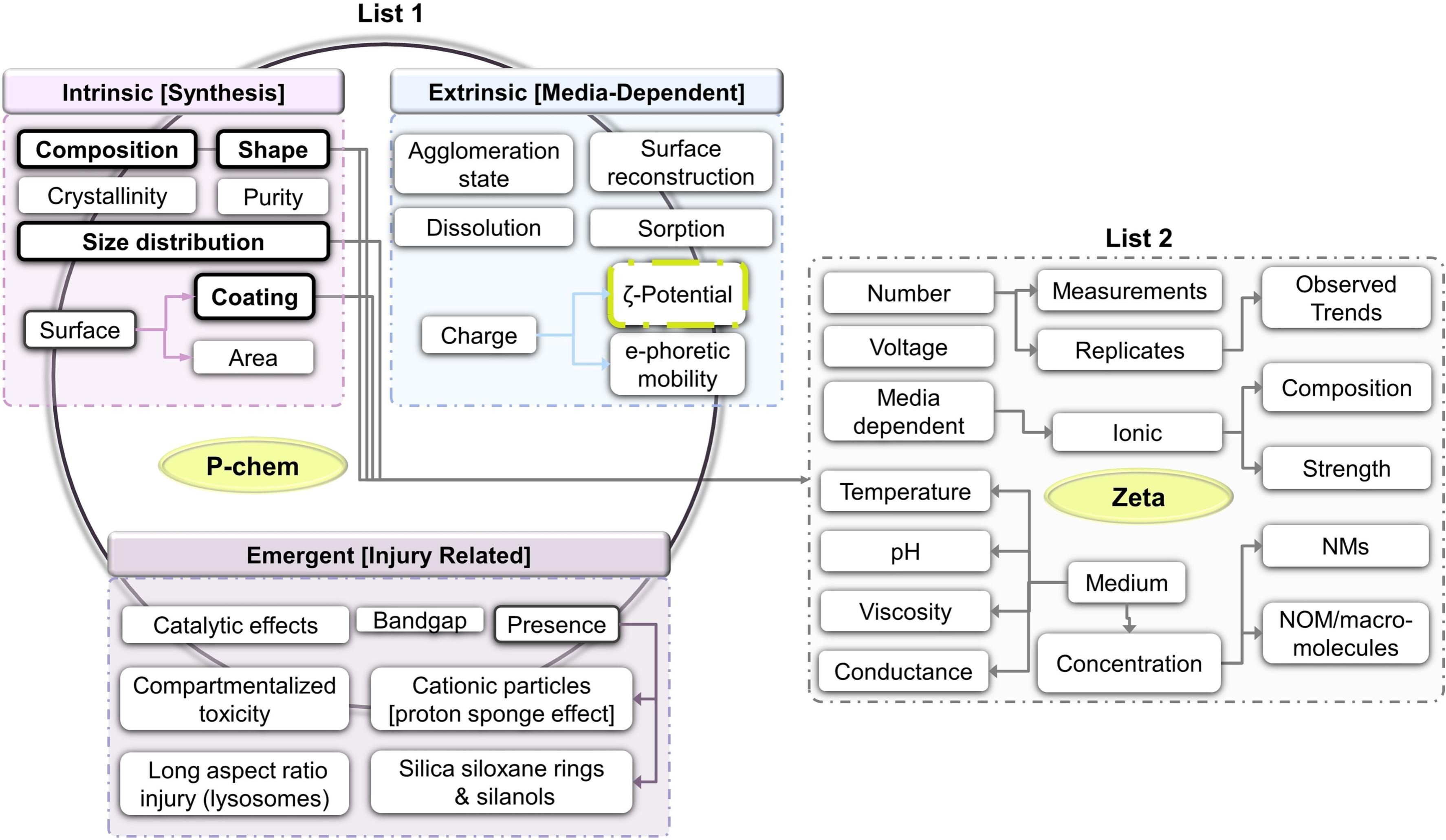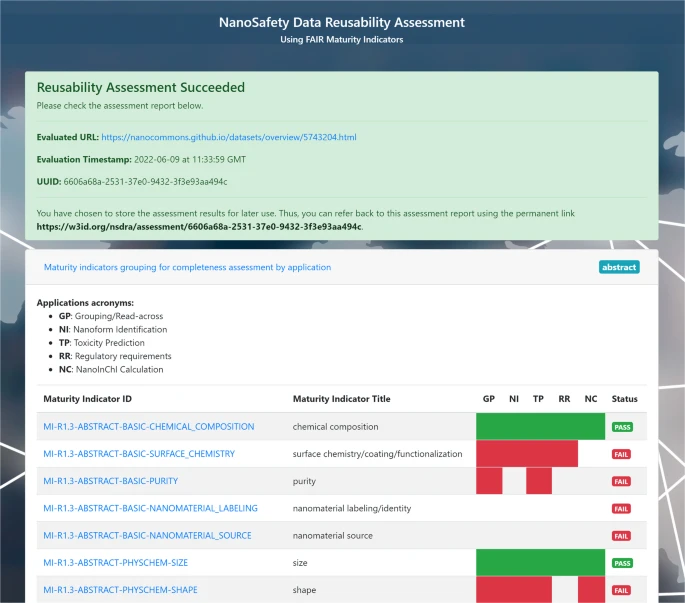
Ammar is finishing up his PhD thesis with his research on the use of FAIR towards predictive toxicology. Or, “AI ready”, as the term FAIR is now sometimes explained. Any computational method needs good data, and just FAIR is not enough. It needs to meet community standards, as formalized in R1.3. To me, this includes meeting community standards like minimal reporting standards.