Last month we released a new version of pdftools and a new companion package qpdf for working with pdf files in R. This release introduces the ability to perform pdf transformations, such as splitting and combining pages from multiple files. Moreover, the pdf_data() function which was introduced in pdftools 2.0 is now available on all major systems.Split and Join PDF files It is now possible to split, join, and compress pdf files with pdftools.
Messages de Rogue Scholar
Version 7.0.0 of drake just arrived on CRAN, and it is faster and easier to use than previous releases.install.packages("drake")Recap Data analysis can be slow. A round of scientific computation can take several minutes, hours, or even days to complete. After it finishes, if you update your code or data, your hard-earned results may no longer be valid. How much of that valuable output can you keep, and how much do you need to update?
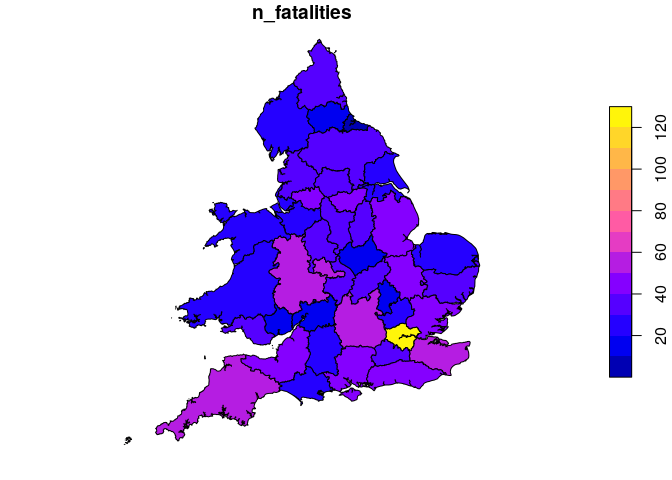
Introduction stats19 is a new R package enabling access to and working withGreat Britain’s official road traffic casualty database,STATS19.
The ssh package provides a native ssh client for R. You can connect to a remote server over SSH to transfer files via SCP, setup a secure tunnel, or run a command or script on the host while streaming stdout and stderr directly to the client. The intro vignette provides a brief introduction.
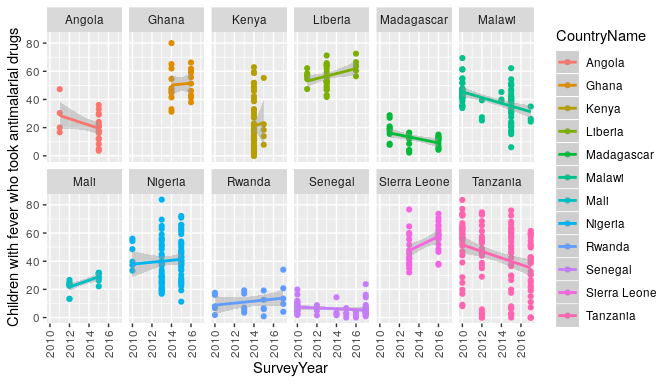
There seem to be a lot of ways to write about your R package, and rather than haveto decide on what to focus on I thought I’d write a little bit about everything.To begin with I thought it best to describe what problem rdhs tries to solve,why it was developed and how I came to be involved in this project.

European eels ( Anguilla anguilla ) have it tough. Not only are they depicted as monsters in movies, they are critically endangered in real life. One of the many aspects that is contributing to their decline is the reduced connectivity between their freshwater and marine habitats.

The Ecology Hackathon Almost one year ago now, ecologists filled a room for the “Ecology Hackathon: Developing R Packages for Accessing, Synthesizing and Analyzing Ecological Data” that was co-organised by rOpenSci Fellow, Nick Golding and Methods in Ecology and Evolution. This hackathon was part of the “Ecology Across Borders” Joint Annual Meeting 2017 of BES, GfÖ, NecoV, and EEF in Ghent.

I never really thought I would write an R package. I use R pretty casually. Then, this year, I was invited to participate during the last week of the Analytical Paleobiology short course, an intensive month-long experience in quantitative paleontology. I was thrilled to be invited.

spatsoc is an R package written by Alec Robitaille, Quinn Webber and Eric Vander Wal of the Wildlife Evolutionary Ecology Lab (WEEL) at Memorial University of Newfoundland. It is the lab’s first R package and was recently accepted through the rOpenSci onboarding process with a big thanks to reviewers Priscilla Minotti and Filipe Teixeira, and editor Lincoln Mullen.
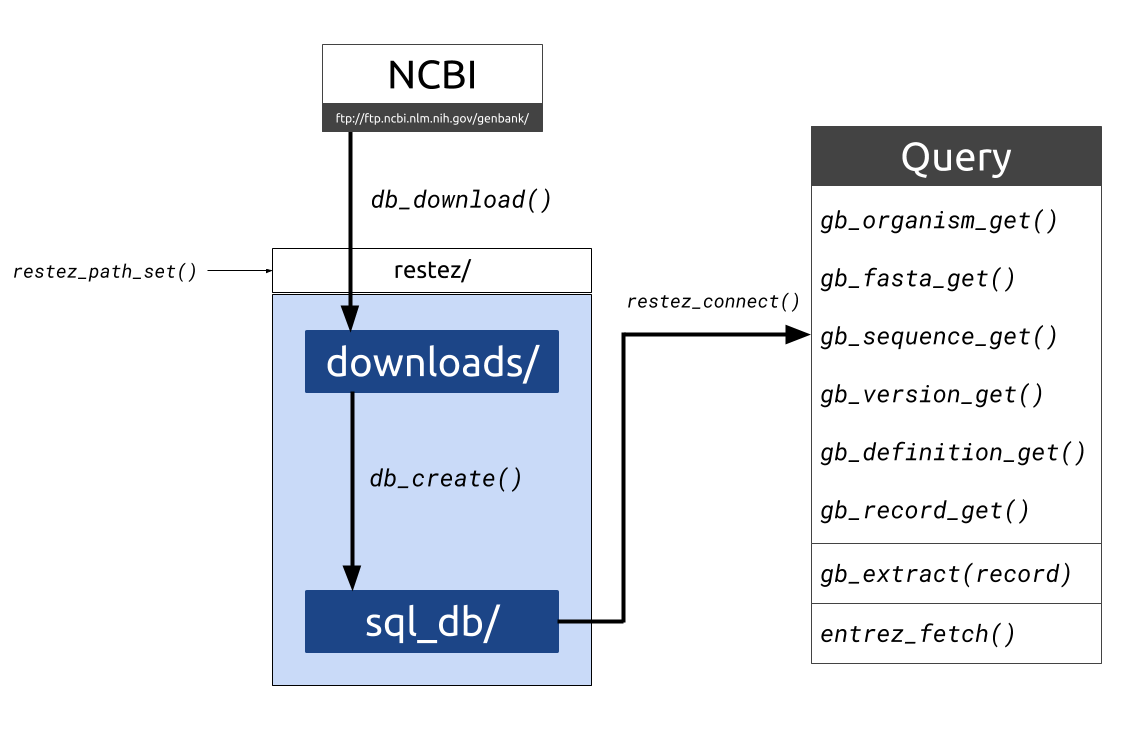
What is restez? R packages for interacting with the National Center for Biotechnology Information (NCBI) have, to-date, depended on API query calls via NCBI’s Entrez.For computational analyses that require the automated look-up of reams of biological sequence data, piecemeal querying via bandwith-limited requests is evidently not ideal.