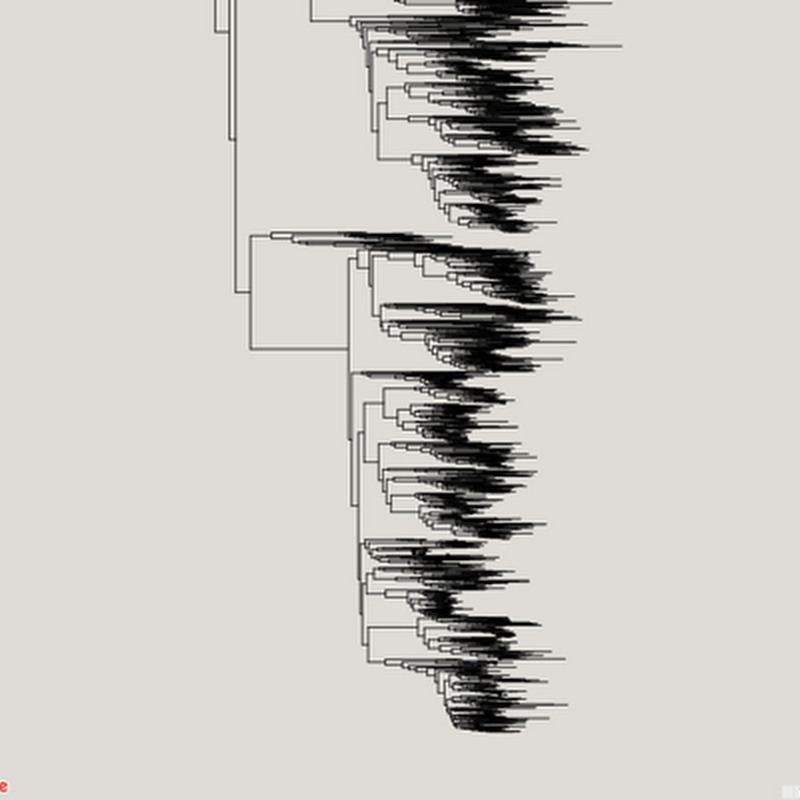
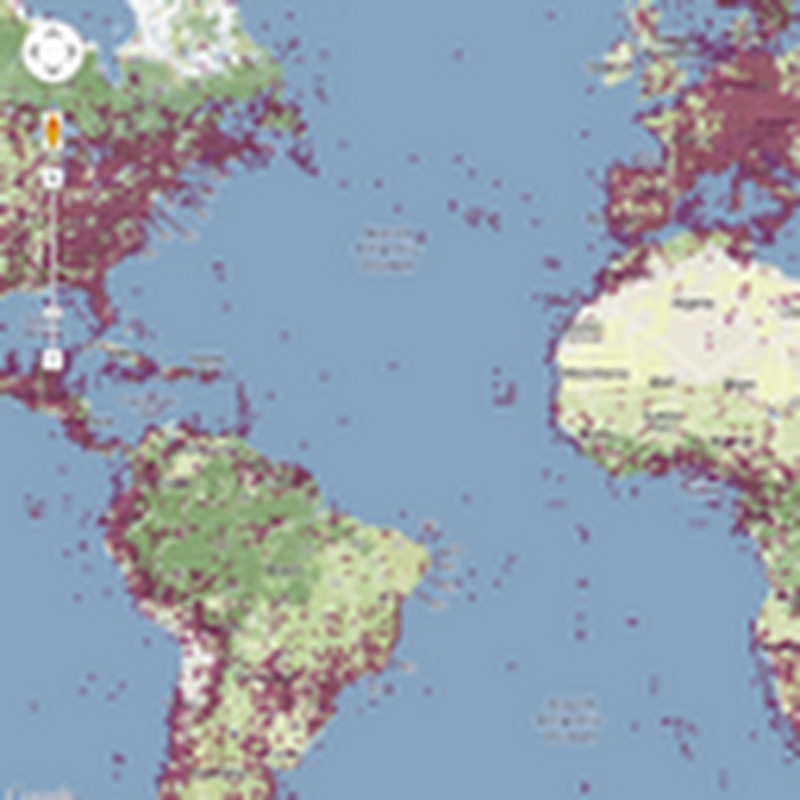
I keep returning to the problem of viewing large graphs and trees, which means my hard drive has accumulated lots of failed prototypes. Inspired by some recent discussions on comparing taxonomic classifications I decided to package one of these (wildly incomplete) prototypes up so that I can document the idea and put the code somewhere safe.