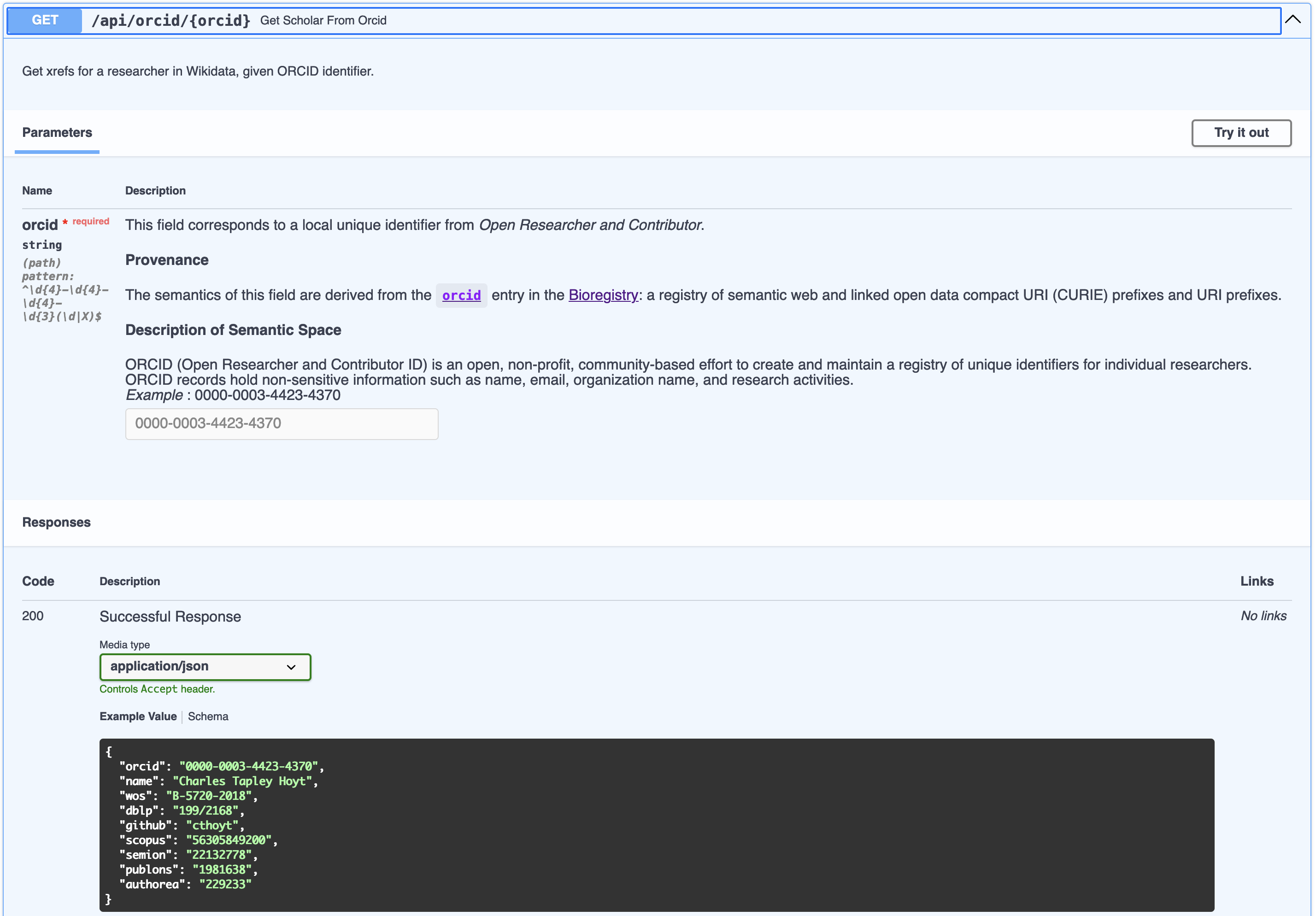
Using Pydantic for encoding data models and FastAPI for implementing APIs on top of them has become a staple for many Python programmers.

Using Pydantic for encoding data models and FastAPI for implementing APIs on top of them has become a staple for many Python programmers.

This week I attended the SWAT4(HC)LS (Semantic Web Applications and Tools for Healthcare and Life Sciences) meeting in Edinburgh. Although a relatively small meeting, SWAT4(HC)LS attracts some big names in the field and featured keynotes by Denny Vrandečić (founder of Wikidata), Dov Greenbaum, Birgitta König-Ries, and Helen Parkinson.

David Attenborough’s latest homage to biodiversity, Blue Planet II is, as always, visually magnificent. Much of its impact derives from the new views of life afforded by technological advances in cameras, drones, diving gear, and submersibles. One might hope that the supporting information online reflected the equivalent technological advances made in describing and sharing information. Sadly, this is not the case.

Continuing my on-again off-again relationship with the Semantic Web, I stumbled across a cool approach to visualising the results of SPARQL queries.
Steve Baskauf has concluded a thoughtful series of blog posts on RDF and biodiversity informatics with http://baskauf.blogspot.co.uk/2015/07/confessions-of-rdf-agnostic-part-7.html. In this post he discussed the "Rod Page Challenge", which was a series of grumpy posts I wrote (starting with this one) where I claimed RDF basically sucked, and to illustrate this I issued a challenge for people to do something interesting with some RDF I provided.
Previously, I had noted that Corey reported in 1963/65 the total synthesis of the sesquiterpene dihydrocostunolide. Compound 16 , known as Eudesma-1,3-dien-6,13-olide was represented as shown below in black; the hydrogen shown in red was implicit in Corey’s representation, as was its stereochemistry. As of this instant, this compound is just one of 64,688,893 molecules recorded by Chemical Abstracts.

I've made some progress on a wiki of phylogenies. Still much to do, but here are some samples of what I'm trying to do. First up, here's an example of a publication http://iphylo.org/treebase/Doi:10.1016/j.ympev.2008.02.021: In addition to basic bibiographic details we have links to GenBank sequences and a phylogeny. The sequences are georeferenced, which enables us to generate a map.
I'm revisiting the idea of building a wiki of phylogenies using Semantic Mediawiki. One problem with a project like this is that it can rapidly explode.
Some months ago now I gave a talk at very exciting symposium organized by Greg Wilson as a closer for the Software Carpentry course he was running at Toronto University.

At the start of this week I took part in a biodiversity informatics workshop at the Naturhistoriska riksmuseets, organised by Kevin Holston. It was a fun experience, and Kevin was a great host, going out of his way to make sure myself and other contributors were looked after. I gave my usual pitch along the lines of "if you're not online you don't exist", and talked about iSpecies, identifiers, and wikis.