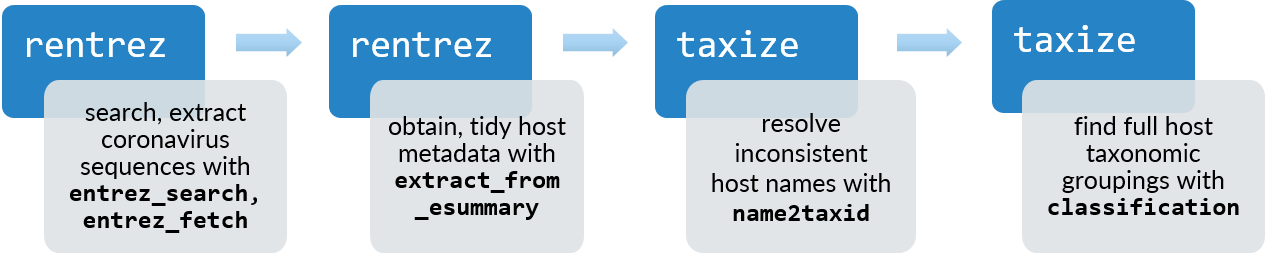
Emerging viruses might be on everyone’s mind right now, but as an epidemiologist and disease ecologist I’ve always been interested in how and why pathogens move from animal hosts to humans.The current pandemic of the disease we call COVID-19 is caused by Severe acute respiratory syndrome (SARS) coronavirus 2 (SARS-CoV-2), a virus that has emerged from wildlife like SARS coronavirus and Middle East respiratory syndrome (MERS) coronavirus