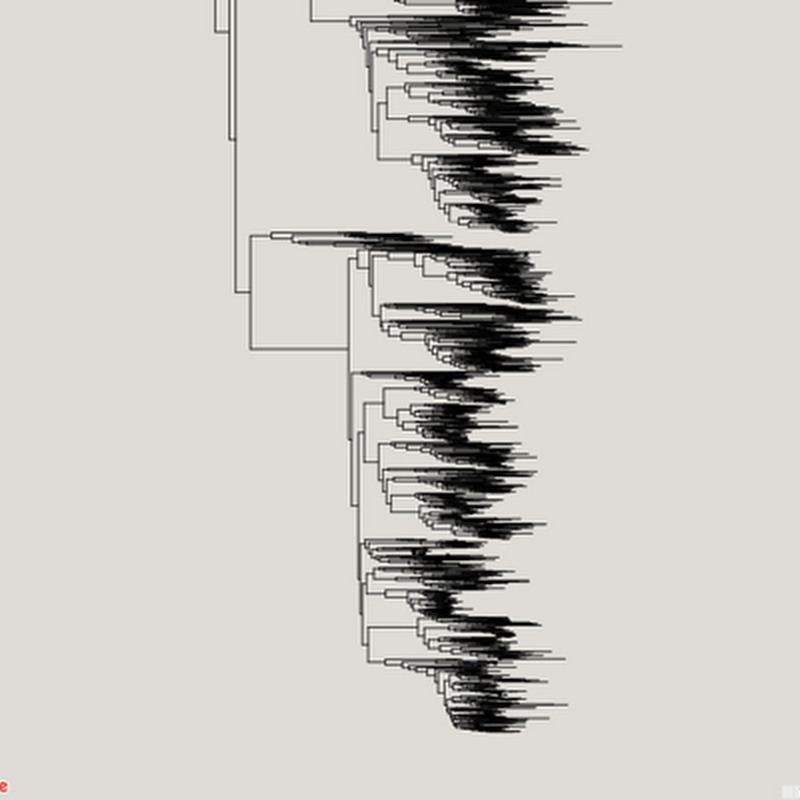
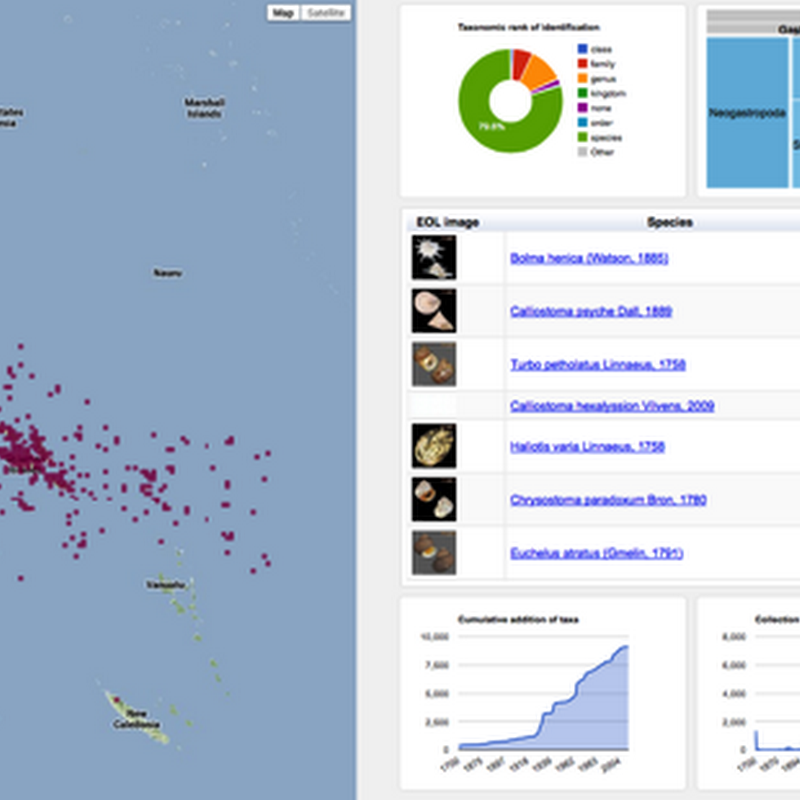
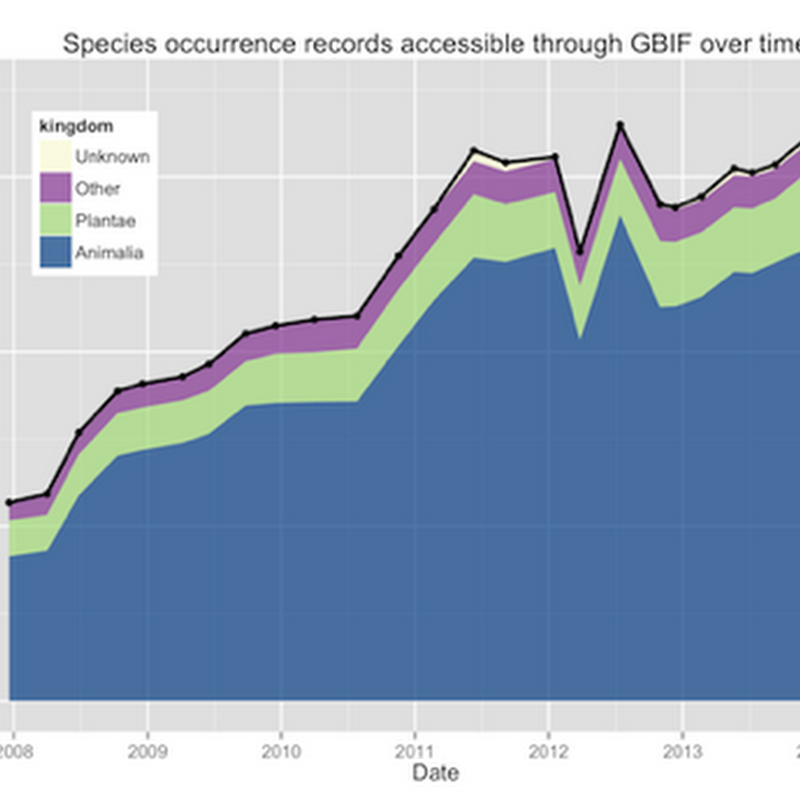
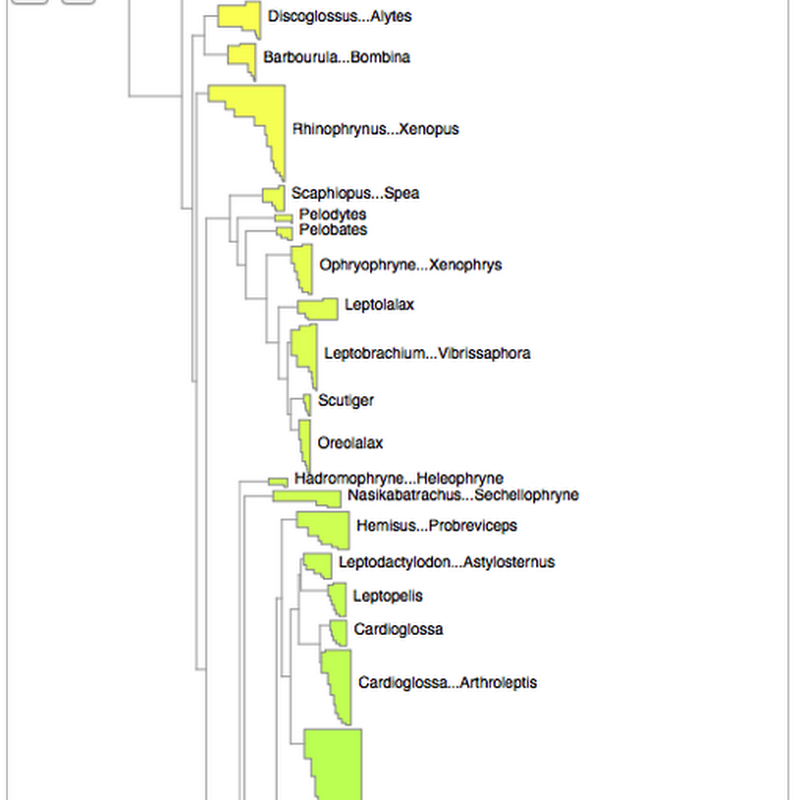
How to cite: Page, R. (2021). Maximum entropy summary trees to display higher classifications https://doi.org/10.59350/af01t-6sw74 A challenge in working with large taxonomic classifications is how you display them to the user, especially if the user probably doesn't want all the gory details.