pubchunks is a package grown out of the fulltext package. fulltextprovides a single interface to many sources of full text scholarly articles. Aspart of the user flow in fulltext there is an extraction step where fulltext::chunks()pulls parts of articles out of XML format article files.
Messaggi di Rogue Scholar
This week an update for xml2 and a new xslt package have appeared on CRAN. A full announcement for xml2 version 1.1 will appear on the rstudio blog. This post explains xml validation (via xsd schema) and xml transformation (via xslt stylesheets) which have been added in this release. XML schemas and stylesheets are not exactly new; both xslt 1.1 (2001) and xsd 1.0 (2004) have been available in browsers for over a decade.
In the beginning (taken here as prior to ~1980) libraries held five-year printed consolidated indices of molecules, organised by formula or name (Chemical abstracts). This could occupy about 2m of shelf space for each five years. And an equivalent set of printed volumes from the Beilstein collection.
One of goals of BioNames is to be more than simply another taxonomic database. In particular, I'm interested in the idea of having a platform for viewing taxonomic publications. One way to think about this is to consider the experience of viewing Wikipedia. For any given page in Wikipedia there will be links to other, related content in Wikipedia.

One of the things I keep revisiting is the way we display scientific articles. Apart from Nature's excellent iPhone and iPad apps, most efforts to re-imagine how we display articles are little more than glorified PDF viewers (e.g., the PLoS iPad app). Part of the challenge is that if we make the article more interactive we immediately confront the problem of how to link to other content.

This post is simply a quick note on some experiments with DjVu that I haven't finished. Much of BHL's content is available as DjVu files, which contain both the scanned images and OCR text, complete with co-ordinates of each piece of text. This means that it would, in principle, be trivial to lay out the bounding boxes of each text element on a web page.

Starting to get serious about the Grand Challenge. First step is to parse the XML data Elsevier made available. Sadly this is only for Molecular Phylogenetics and Evolution for 2007, I would have liked the whole journal in XML to avoid hassles with parsing PDF. However, XML is not without it's own problems.
Yesterday I installed the Eclipse Web Tools Platform again, and now succesfully, using the Eclipse update mechanism, on my Kubuntu dapper eclipse install. Because it has a validating XML editor, the one last thing I still needed jEdit for. (I do miss the vertical selection feature of jEdit, though.) It signals me of errors, and allows autocompletion.
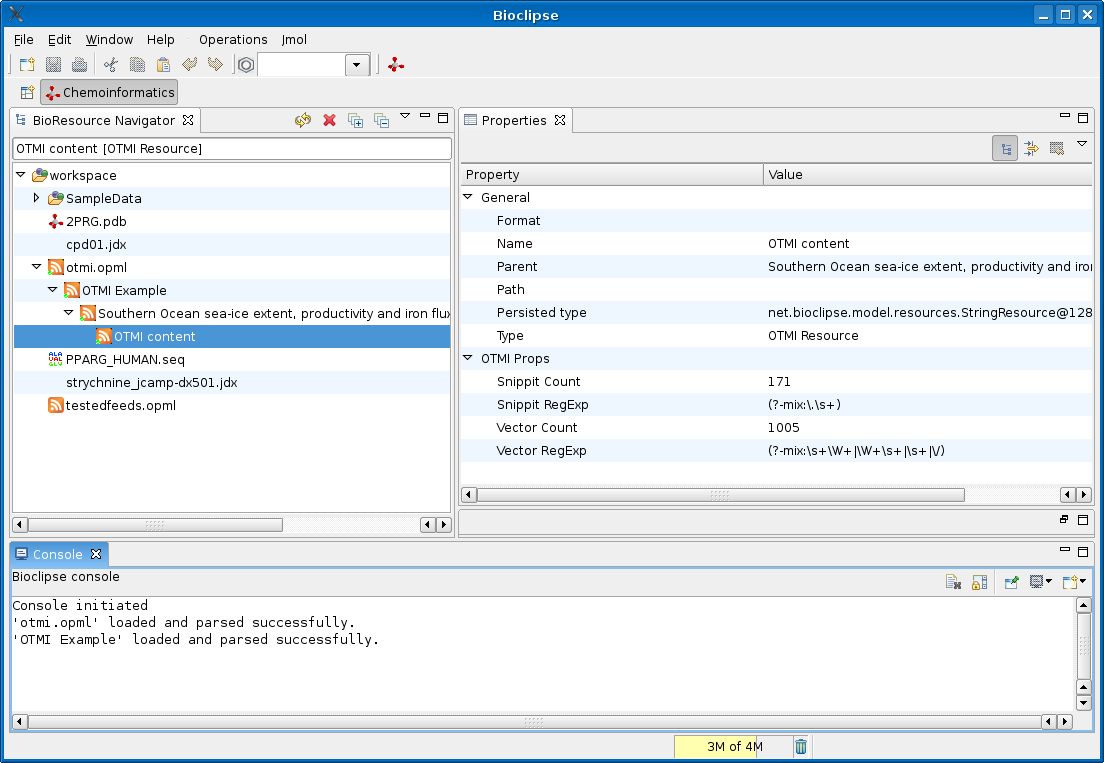
Timo Hannay blogged in Nature’s Nascent blog about the Open Text Mining Interface (OTMI), which is “a suggestion from Nature about how we might achieve text-mining and indexing purposes”. The idea is that each article has a link pointing to a machine readable file containing raw data about (and from?) the article.