Dealing with taxonomic inconsistencies within and across datasets is a fundamental challenge of ecology and evolutionary biology. Accounting for species synonyms, taxa splitting and unification is especially important as aggregation of data across time and different data sources becomes increasingly common.
Messaggi di Rogue Scholar

It was the best of times, it was the worst of times. Dickens might have meant it figuratively, but in the case of the rOpenSci OzUnconf 2019, we mean it literally. Set to the backdrop of a national emergency that is still ongoing from 11-13 December, our participants came from across Australia as well as New Zealand, Japan, India and Indonesia.
babette 1 is a package to work with BEAST2 2 ,a software platform for Bayesian evolutionary analysis from R. babette is a spin-off of my own academic research.As a PhD I work on models of diversification: mathematical descriptionsof how species form new species.
In January 2019, we announced the release of rOpenSci’s Code of Conduct version 2.0. This includes a named Committee, greater detail about unacceptable behaviors, instructions on how to make a report, and information on how reports are handled. We are committed to transparency with our community while upholding of victims and people who report incidents.
One year ago, we released our new Code of Conduct. At that time, the Code of Conduct Committee (authors of this post) agreed to do an annual review of the text, reporting form, and our internal guidelines. We have made one change to the Code of Conduct text.

We mean it. On behalf of rOpenSci, thank you to everyone who has contributed their creativity, curiosity, smarts, and time in the last year. We are fortunate to have paid staff who work to build technical and social infrastructure to lower barriers to working with research data. But it is our community, built on trust, that binds us together and helps us see who we are working for.
Want to get some hands-on insights into running an open source community? Here’s an opportunity to work with me, rOpenSci’s Community Manager, on some non-code community-related work. I am looking for someone to work 1 day a week for 12 to 14 weeks. Working alongside rOpenSci’s Community Manager, Stefanie Butland, you will use guidelines and checklists to help run some of our established programs like our Blog and Community Calls.

We’re thrilled to be introducing a new member of our team. Mark Padgham has joined rOpenSci as a Software Research Scientist working full-time from Münster, Germany. Mark will play a key role in research and development of statistical software standards and expanding our efforts in software peer review, enabled by new funding from the Sloan Foundation.
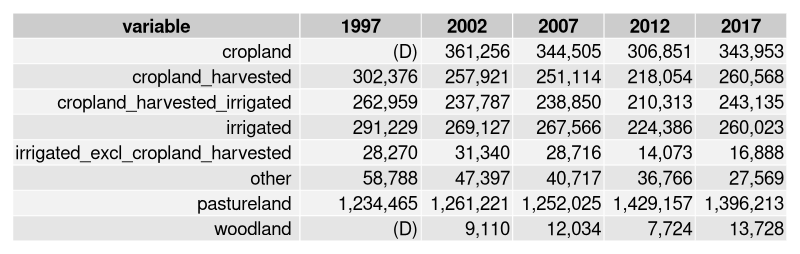
The United States Deparment of Agriculture National AgriculturalStatistics Service (USDA-NASS) provides a wide range of agriculturaldata that includes animal, crop, demographic, economic, andenvironmental measures across a number of geographies and time periods.This data is available by direct download or queriable via theQuick Stats interface.

rOpenSci thrives because of volunteer contributions from community members - submitting and reviewing R packages, serving as editors for software peer review, writing blog posts, sharing information about packages and resources, contributing code and documentation and answering others’ questions. Recently our fiscal sponsor, NumFOCUS, gave us an opportunity to nominate two contributors for recognition at the NumFOCUS annual summit.