The Apache Tika parser is like the Babel fish in Douglas Adam’s book, “The Hitchhikers’ Guide to the Galaxy” 1 . The Babel fish translates any natural language to any other. Although Tika does not yet translate natural language, it starts to tame the tower of babel of digital document formats. As the Babel fish allowed a person to understand Vogon poetry, Tika allows an analyst to extract text and objects from Microsoft Word.
Messaggi di Rogue Scholar
library(tidyverse)library(monkeylearn) This is a story (mostly) about how I started contributing to the rOpenSci package monkeylearn. I can’t promise any life flipturning upside down, but there will be a small discussion about git best practices which is almost as good 🤓. The tl;dr here is nothing novel but is something I wish I’d experienced firsthand sooner.
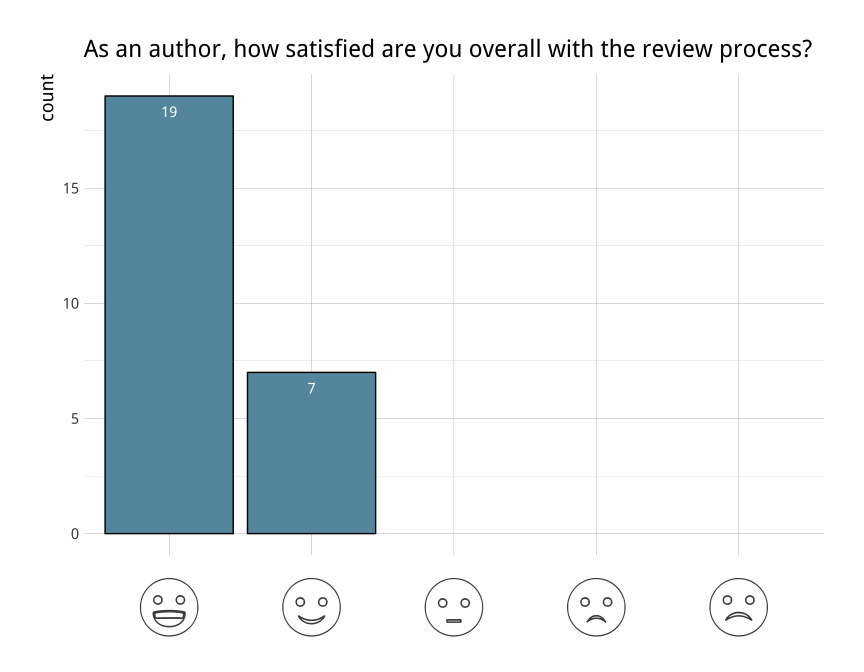
rOpenSci’s package review system (akaonboarding) is one of our keyactivities to improve quality and sustainability of scientific Rpackages. Theeditorial team are constantly working towards improving the experiencefor both authors and reviewers. After our first year, we surveyedauthors andreviewers whoparticipated in our onboarding process to help us better understandwhat’s working well and where there is room for improvement.

KO: What is your name, job title, and how long have you been using R? NR: I’m Noam Ross, I’m a Senior Research Scientist at EcoHealth Alliance, a non-profit that works at the intersection of conservation and health. I’ve done work in R for about 8 years, which is essentially from the start of graduate school (five years) and three years of work. KO: Did you love it [R] immediately? NR: No! And I’m mixed on whether I love it now.
The general struggle Something that will make life easier in the long-run can be the most difficult thing to do today . For coders, prioritising the long term may involve an overhaul of current practice and the learning of a new skill.
If you read my reflection #1 on rOpenSci Onboarding, then you know I see value in the Onboarding process. A LOT of value even. This post is about where that value lies. This question has important corollaries which I will explore here based on my experience as a reviewer of bowerbird: How is a package peer reviewer’s time best spent? When is the best time in a software package’s life cycle to undertake peer review?
That’s a lot alike Data Science, isn’t it? Hydrologic Processes evolve in space and time, are extremely complex and we may never comprehend them. For this reason Hydrologists use models where their inputs and outputs are measurable variables: climatic and hydrologic data, land uses, vegetation coverage, soil type etc.
Olfactory Coding Detecting volatile chemicals and encoding these into neuronal activity is a vital task for all animals that is performed by their olfactory sensory systems. While these olfactory systems vary vastly between species regarding their numerical complexity, they are amazingly similar in their general structure.
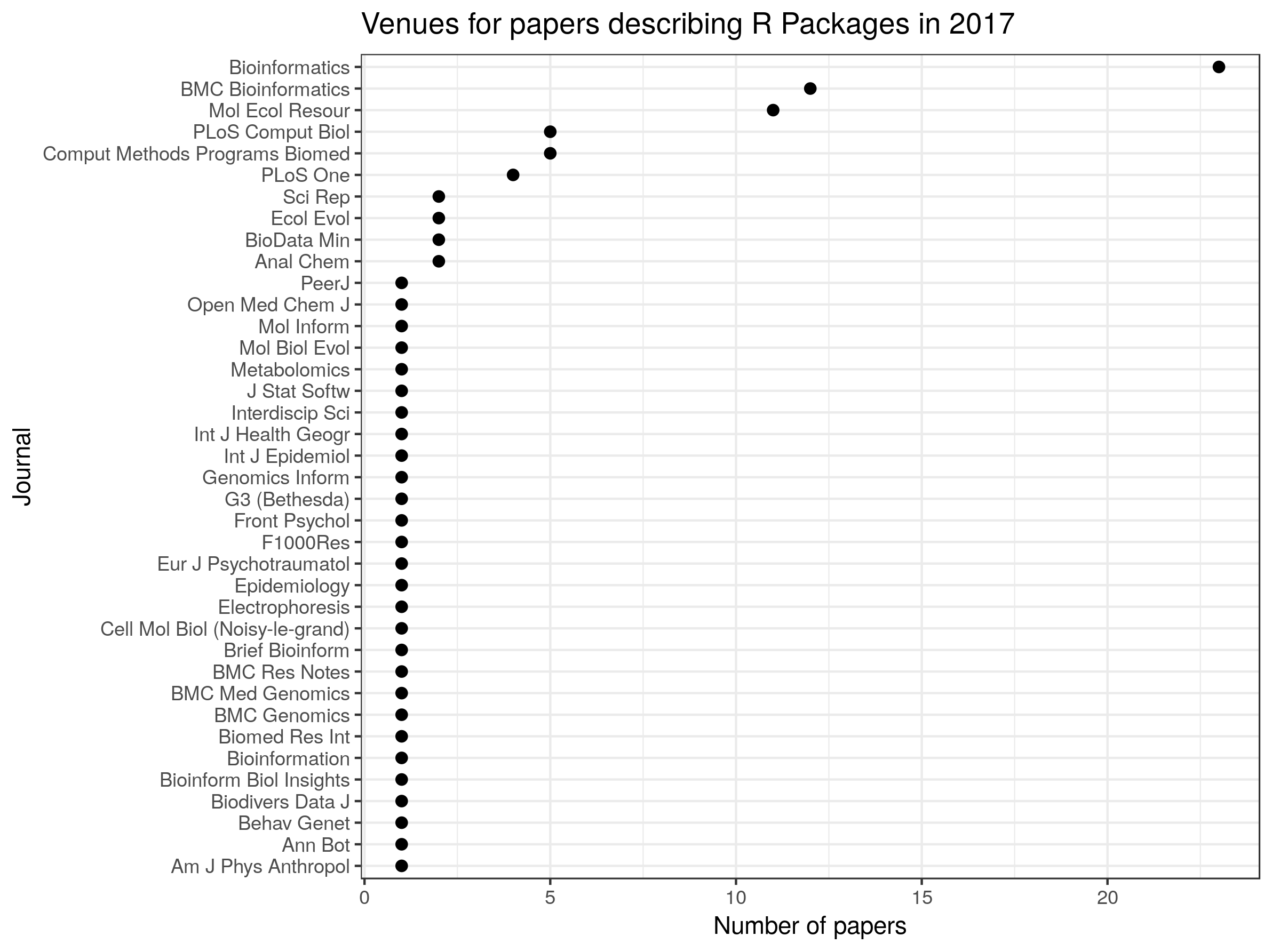
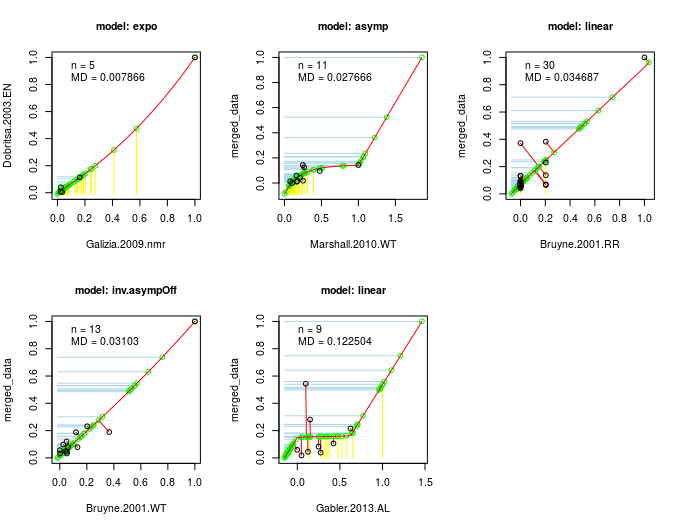
I am happy to say that the latest issue of The R Journal includes a paperdescribing rentrez,the rOpenSci package for retrieving data from the National Center for Biotechnology Information(NCBI). The NCBI is one of the most important sources of biological data. The centreprovides access to information on 28 million scholarly articles through PubMed and 250million DNA sequences through GenBank.
At rOpenSci, our R package peer review process relies on the the hard work of many volunteer reviewers. These community members donate their time and expertise to improving the quality of rOpenSci packages and helping drive best practices into scientific software. Our open review process, where reviews and reviewers are public, means that one benefit for reviewers is that they can get credit for their reviews.