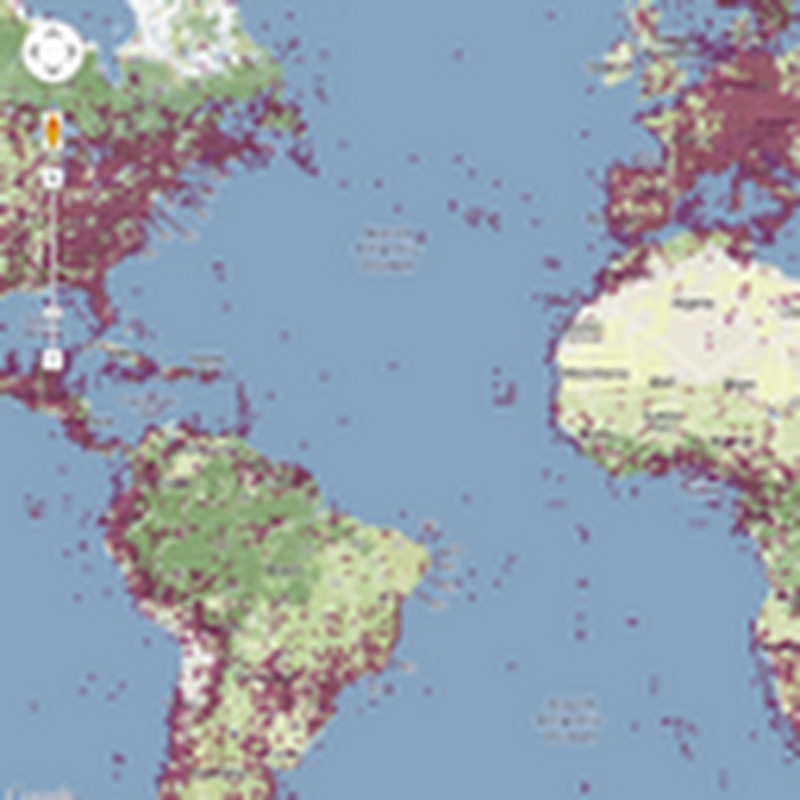
Some notes to self about future directions for the "million DNA barcodes map" http://iphylo.org/~rpage/bold-map/. At the moment we have an interactive map that we can pan and zoom, and click on a marker to get a list of one or more barcodes at the location. We can also filter by major taxonomic group. Here are some ideas on what could be next. Search At the moment search is simply browsing the map.