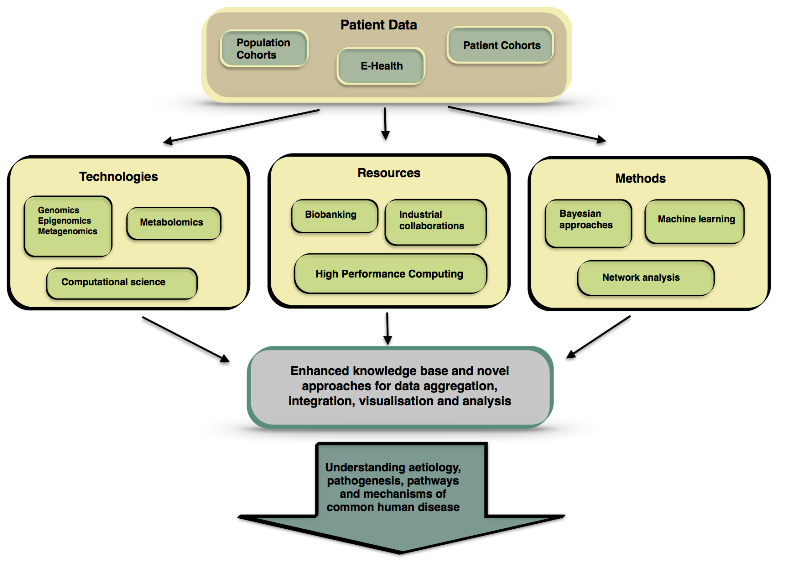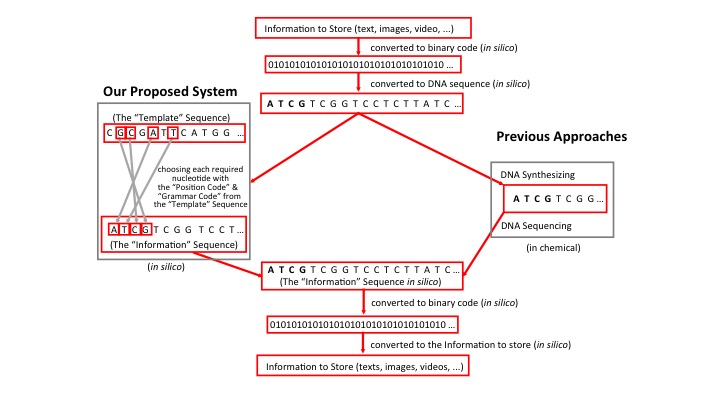
With the current annual data creation rate estimated to be in the tens of zettabytes, the flood of information currently being generated in every area of human life is crashing up against limited data storage solutions. However, DNA, which serves as a storage system for biological information, has been proposed as a potential means to store an unlimited amount of information.