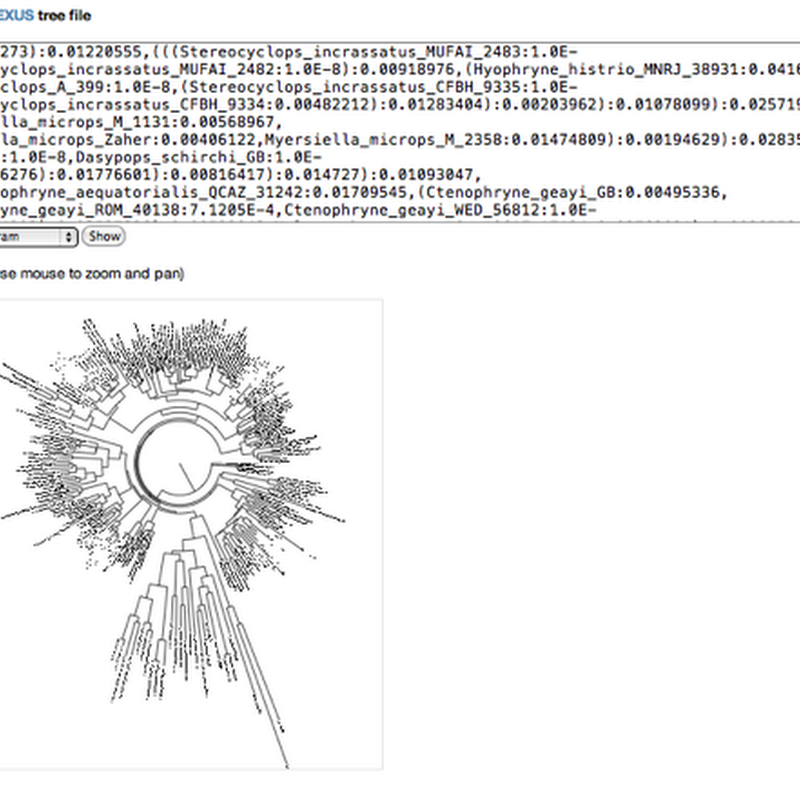
Following on from the SVG experiments I've started to put some of the Javascript code for displaying phylogenies on Github. Not a repository yet, but as gists, little snippets of code. Mike Bostock has created http://bl.ocks.org/ which makes it possible to host gists as working examples, so you can play with the code "live". The first gist takes a Newick tree, parses it and displays a tree.


