
I've added an experimental feature to BioStor that uses data from Wikidata and Wikispecies to augment what information BioStor displays on authors.

I've added an experimental feature to BioStor that uses data from Wikidata and Wikispecies to augment what information BioStor displays on authors.

[We are excited to announce the launch of the Europe PMC author profile pages! Based on your ORCID record, they provide a graphical overview of your publications in Europe PMC and your citation rate over time.]{style=“background-color: transparent; color: #666666; font-family: "arial"; font-size: 14.66px; font-style: normal; font-variant: normal; font-weight: 400; text-decoration: none;

One of the less glamorous but necessary tasks of data cleaning is mapping "strings to things", that is, taking strings such as "George A. Boulenger" and mapping them to identifiers, such as ISNI: 0000 0001 0888 841X. In case of authors such as George Boulenger, one way to do this would be through Wikipedia, which has entries for many scientists, often linked to identifiers for those people (see the bottom of the Wikipedia page for George A.

[Title quote from ‘Sonnets from the Portuguese’, Sonnet 43, by Elizabeth Barrett Browning.]{style=“font-family: "helvetica neue" , "arial" , "helvetica" , sans-serif;”} [Our relationship with ORCID personal identifiers blooms in a number of ways:]{style=“font-family: "helvetica neue" , "arial" , "helvetica" , sans-serif;”} [ ]{style=“font-family: "helvetica neue" , "arial" , "helvetica" , sans-serif;”} [ ]{style=“font-family:

I spent last Friday and Saturday at ( Research in the 21st Century: Data, Analytics and Impact , hashtag #ReCon_15) in Edinburgh. Friday 19th was conference day, followed by a hackday at CodeBase. There's a Storify archive of the tweets so you can get a sense of the meeting. Sitting in the audience a few things struck me. No identifier wars, DOIs have won and are everywhere.
Geoff Bilder, Jennifer Lin, Cameron Neylon The announcement of a $3M grant from the Helmsley Trust to ORCID is a cause for celebration.
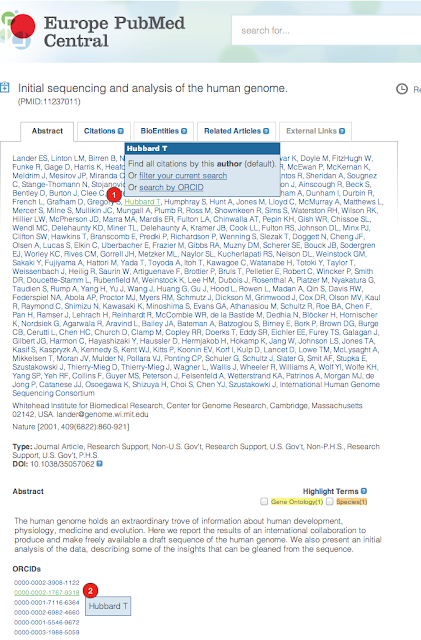
[Following the development of the ORCID-based Article Claiming Tool (see this blog post), Europe PMC has now integrated ORCIDs into its website, search systems, and web services.

This is a quick sketch of a way to combine existing tools to help clean and annotate data in GBIF, particularly (but not exclusively) occurrence data. GitHub The data provider puts a Darwin Core Archive (expanded, not zipped) into a GitHub repository. GBIF forks the repository, cleans the data, and uploads that to GBIF to populate the database behind the portal.
[Europe PMC is pleased to offer researchers the ability to link articles to an ORCID. The tool can also be found under the ‘Resources’ menu on the Europe PMC home page.