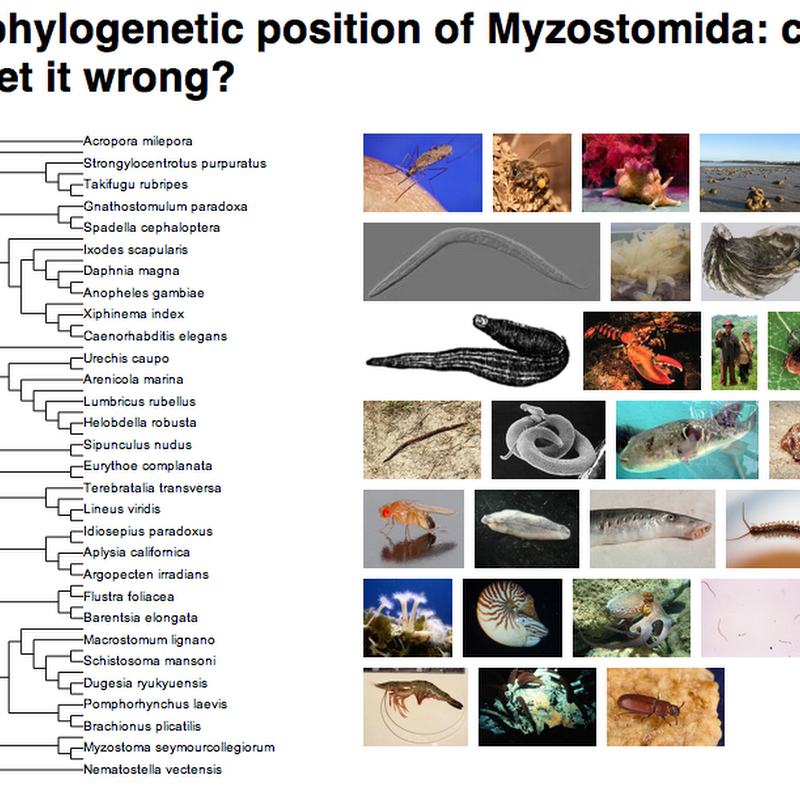
Prompted by a conversation with Vince Smith at the recent Online Taxonomy meeting at the Linnean Society in London I've been revisiting touch-based displays of large trees. There are a couple of really impressive examples of what can be done. Perceptive Pixel I've blogged about this before, but came across another video that better captures the excitement of touch-based navigation of a taxonomy.