
I spend a lot of time searching the web for bibliographic metadata and links to digitised versions of publications.

I spend a lot of time searching the web for bibliographic metadata and links to digitised versions of publications.

Google's Knowledge Graph can enhance search results by display some structured information about a hit in your list of results. It's available in the US (i.e., you need to use www.google.com, although I have seen it occasionally appear for google.co.uk. Here is what Google displays for Eidolon helvum (the straw-coloured fruit bat). You get a snippet of text from Wikipedia, and also a map from the BBC Nature Wildlife site.
[Are you interested in finding highly cited articles? If so then the new features in UKPMC will be of value to you. Search results can now be sorted by the number of times articles have been cited.]{style=“font-family: "trebuchet ms" , sans-serif;”} [Each search result is now shown with a count of the number of articles citing that record.

Prompted by the appearance on the BHL blog of an article about BioStor I've thinking about how to improve what is basically a fairly clunky tool. One major weakness is searching the collection of nearly 40,000 articles extracted from BHL. Note the word "extracted." BioStor isn't a tool like PubMed or Google Scholar where the goal is to find articles on a topic.
Jeff Atwood, one of the co-founders of Stack Overflow recently wrote a blog post Trouble In the House of Google, where he noted that several sites that scrape Stack Overflow content (which Stack Overflow's CC-BY-SA license permits) appear higher in Google's search rankings than the original Stack Overflow pages . When Stack Overflow chose the CC-BY-SA license they made the assumption that: Jeff Atwood's post goes on to argue
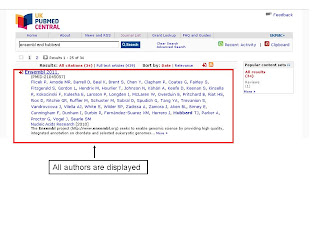
In response to user feedback, the UKPMC search results page now displays all the authors associated with any published article. Prior to this change, the search results page had shown just the first seven authors, with the full set of authors only being visible from the abstract and full-text views. Further feedback on the UKPMC service is welcomed.
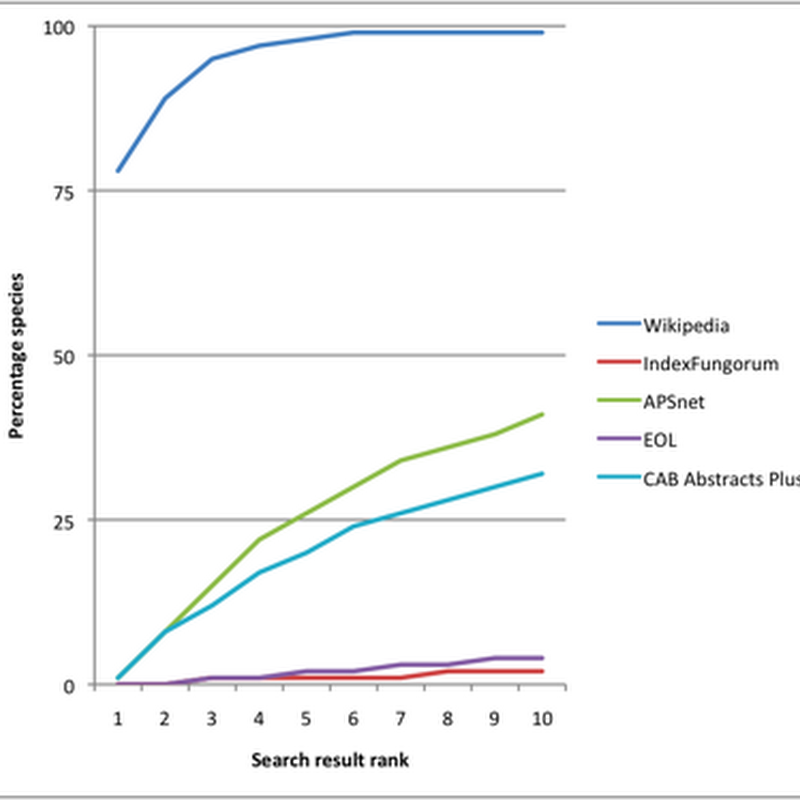
One response to the analysis I did of the Google rank of mammal pages in Wikipedia is to suggest that Wikipedia does well for mammals because these are charismatic. It's been suggested that for other groups of taxa Wikipedia might not be so prominent in the search results. As a quick test I extracted the 1552 fungal species I could find in Wikipedia and repeated the analysis.

One assumption I've been making so far is that when people search for information on an organism using its scientific name, Wikipedia will dominate the search results (see my earlier post for an example of this assumption). I've decided to quantify this by doing a little experiment. I grabbed the Mammal Species of the World taxonomy and extracted the 5416 species names. I then used Google's AJAX search API to look up each name in Google.
Simply click this button to add the UKPMC search plugin to your browser search toolbar. Supported by Firefox 2+ and Internet Explorer 7 only.

Rutger Vos asked on Twitter "What would people want/expect from taxon searching on TreeBASE?". This is a good question, and one which motivated the work I did on TBMap (see doi:10.1186/1471-2105-8-158), which developed a mapping between TreeBASE taxa and other databases. In that paper I published a table showing the effectiveness of string and hierarchical queries of TreeBASE.