
As part of a project exploring GBIF data I've been playing with displaying GBIF data on Google Maps.

As part of a project exploring GBIF data I've been playing with displaying GBIF data on Google Maps.

VertNet has announced that they have implemented issue tracking using GitHub. This is a really interesting development, as figuring out how to capture and make use of annotations in biodiversity databases is a problem that's attracting a lot of attention.

Quick notes on yet another attempt to marry the task of editing a taxonomic classification with versioning it in GitHub. The idea of dumping the whole GBIF classification into GitHub as a series of nested folders looks untenable. So, maybe there's another way to tackle the problem.
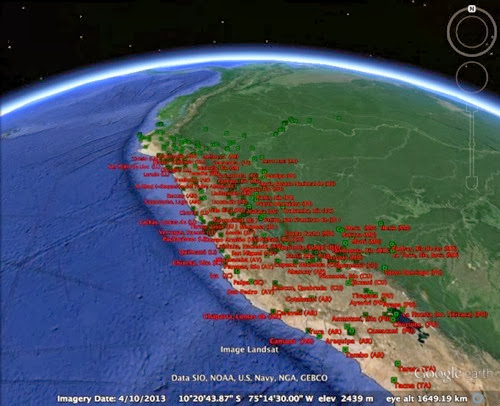
Here's another example of a Darwin Core Archive that is "broken" such that GBIF is missing some information. GBIF data set A checklist to the wasps of Peru (Hymenoptera, Aculeata) comes from Pensoft, and corresponds to the paper: As with the previous example GBIF says there are 0 georeferenced records in this dataset. This is odd, because the ZooKeys page for this article lists three supplementary files, including KML files for Google Earth.

Following on from Annotating and cleaning GBIF data: Darwin Core Archive, GitHub, ORCID, and DataCite here's a quick and dirty example of using GitHub to help clean up a Darwin Core Archive. The dataset 3i - Cicadellinae Database has 2,152 species and 4,749 taxa, but GBIF says it has no georeferenced data.

This is a quick sketch of a way to combine existing tools to help clean and annotate data in GBIF, particularly (but not exclusively) occurrence data. GitHub The data provider puts a Darwin Core Archive (expanded, not zipped) into a GitHub repository. GBIF forks the repository, cleans the data, and uploads that to GBIF to populate the database behind the portal.

In a previous post (Learning from eLife: GitHub as an article repository) I discussed the advantages of an Open Access journal putting its article XML in a version-controlled repository like GitHub. In response to that post Pensoft (the publisher of ZooKeys ) did exactly that, and the XML is available at https://github.com/pensoft/ZooKeys-xml. OK, "now what?" I hear you ask.
Playing with my eLife Lens-inspired article viewer and some recent articles from ZooKeys I regularly come across articles that are incorrectly marked up. As a quick reminder, my viewer takes the DOI for a ZooKeys article (just append it to http://bionames.org/labs/zookeys-viewer/?doi=, e.g. http://bionames.org/labs/zookeys-viewer/?doi=10.3897/zookeys.316.5132), fetches the corresponding XML and displays the article.
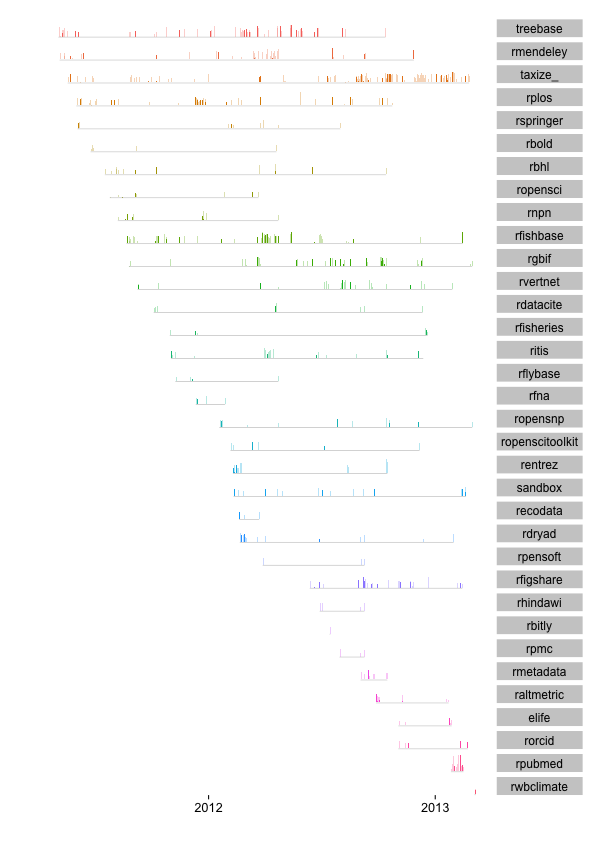
We have been writing code for R packages for a couple years, so it is time to take a look back at the data. What data you ask? The commits data from GitHub ~ data that records who did what and when. Using the Github commits API we can gather data on who commited code to a Github repository, and when they did it. Then we can visualize this hitorical record.

For a current project I'm currently working I show organism distributions using data from GBIF, and I display that data on a map that uses the equirectangular projection.