
Stefanie Butland, rOpenSci Community Manager Some things are just irresistible to a community manager – PhD student Hugo Gruson’s recent tweets definitely fall into that category.

Stefanie Butland, rOpenSci Community Manager Some things are just irresistible to a community manager – PhD student Hugo Gruson’s recent tweets definitely fall into that category.
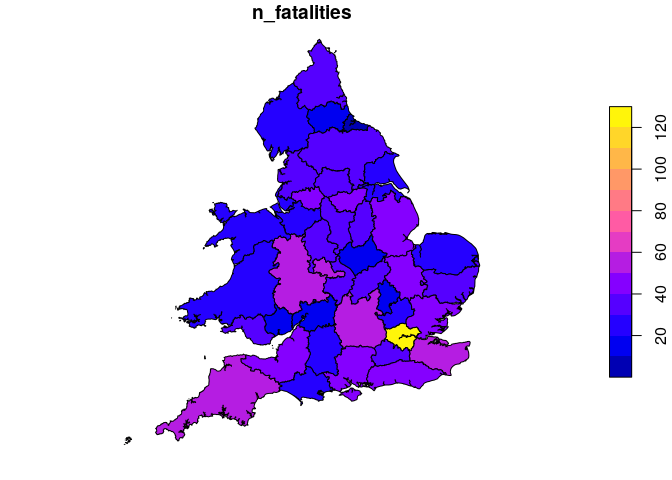
Introduction stats19 is a new R package enabling access to and working withGreat Britain’s official road traffic casualty database,STATS19. We started the package in late 2018 following three main motivations: The release of the 2017 road crash statistics, which showedworsening road safety in some areas, increasing the importance ofmaking the data more accessible.

rOpenSci’s suite of packages is comprised of contributions from staff engineers and the wider R community, bringing considerable diversity of skills, expertise and experience to bear on the suite. How do we ensure that every package is held to a high standard?

We are pleased to welcome Brooke Anderson and Melina Vidoni to our team of Associate Editors for rOpenSci Software Peer Review. They join Scott Chamberlain, Anna Krystalli, Lincoln Mullen, Karthik Ram, Noam Ross and Maëlle Salmon. With the addition of Brooke and Melina, our editorial board now includes four women and four men, located in North America, South America and Europe.
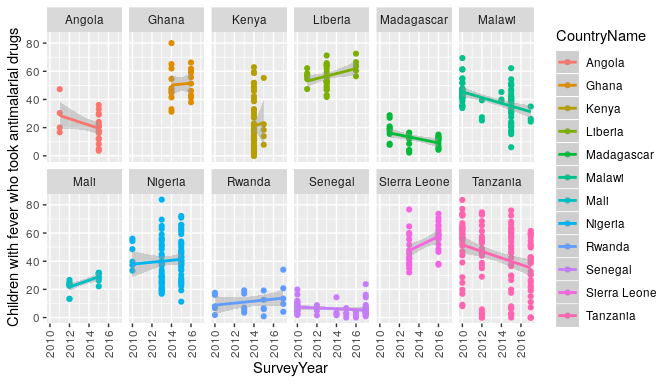
There seem to be a lot of ways to write about your R package, and rather than haveto decide on what to focus on I thought I’d write a little bit about everything.To begin with I thought it best to describe what problem rdhs tries to solve,why it was developed and how I came to be involved in this project.

European eels ( Anguilla anguilla ) have it tough. Not only are they depicted as monsters in movies, they are critically endangered in real life. One of the many aspects that is contributing to their decline is the reduced connectivity between their freshwater and marine habitats.

The Ecology Hackathon Almost one year ago now, ecologists filled a room for the “Ecology Hackathon: Developing R Packages for Accessing, Synthesizing and Analyzing Ecological Data” that was co-organised by rOpenSci Fellow, Nick Golding and Methods in Ecology and Evolution. This hackathon was part of the “Ecology Across Borders” Joint Annual Meeting 2017 of BES, GfÖ, NecoV, and EEF in Ghent.

I never really thought I would write an R package. I use R pretty casually. Then, this year, I was invited to participate during the last week of the Analytical Paleobiology short course, an intensive month-long experience in quantitative paleontology. I was thrilled to be invited.
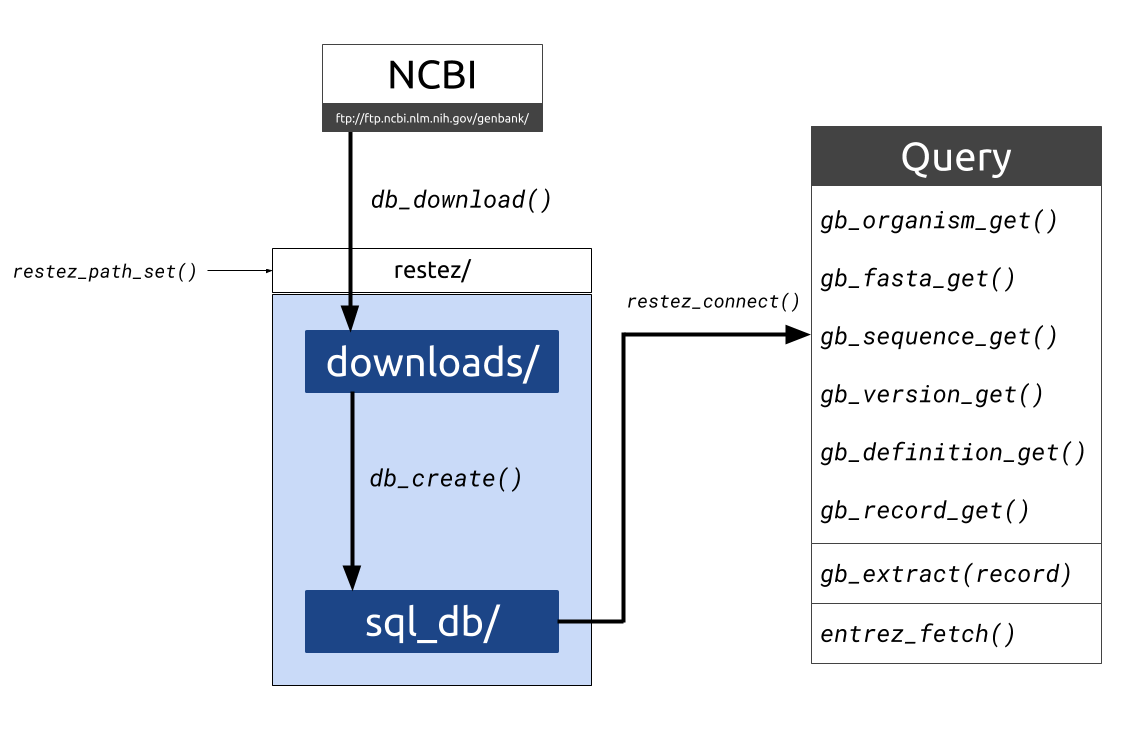
What is restez? R packages for interacting with the National Center for Biotechnology Information (NCBI) have, to-date, depended on API query calls via NCBI’s Entrez.For computational analyses that require the automated look-up of reams of biological sequence data, piecemeal querying via bandwith-limited requests is evidently not ideal.
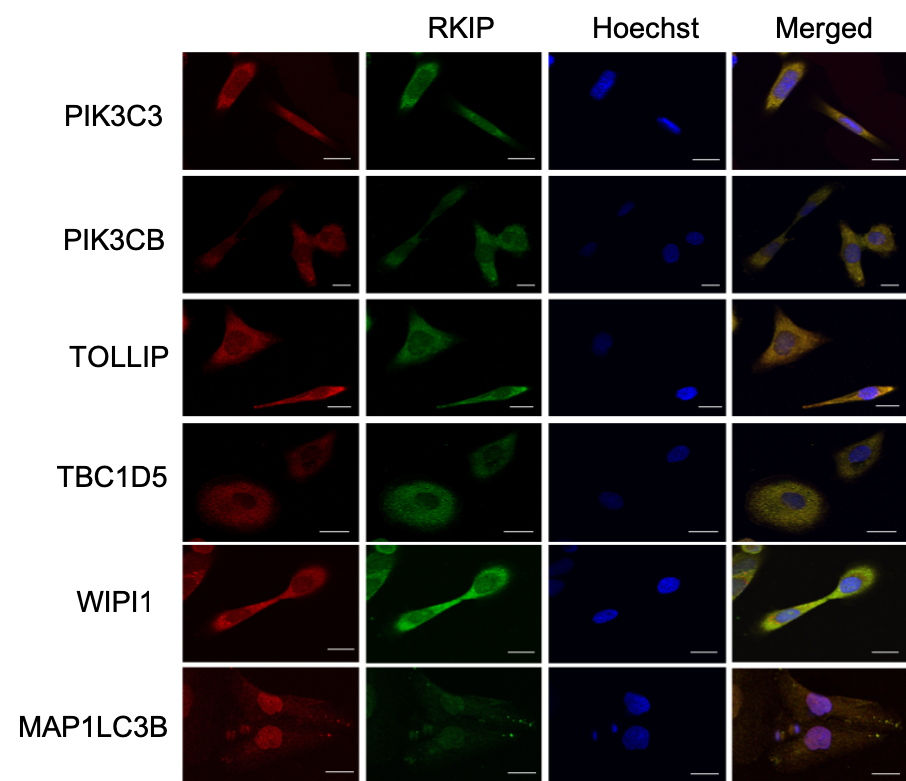
A few months ago, I wasn’t sure what to expect when looking at fluorescence microscopy images in published papers. I looked at the accompanying graph to understand the data or the point the authors were trying to make. Often, the graph represents one or more measures of the so-called co-localization, but I couldn’t figure out how to interpret them. It turned out; reading the images is simple.