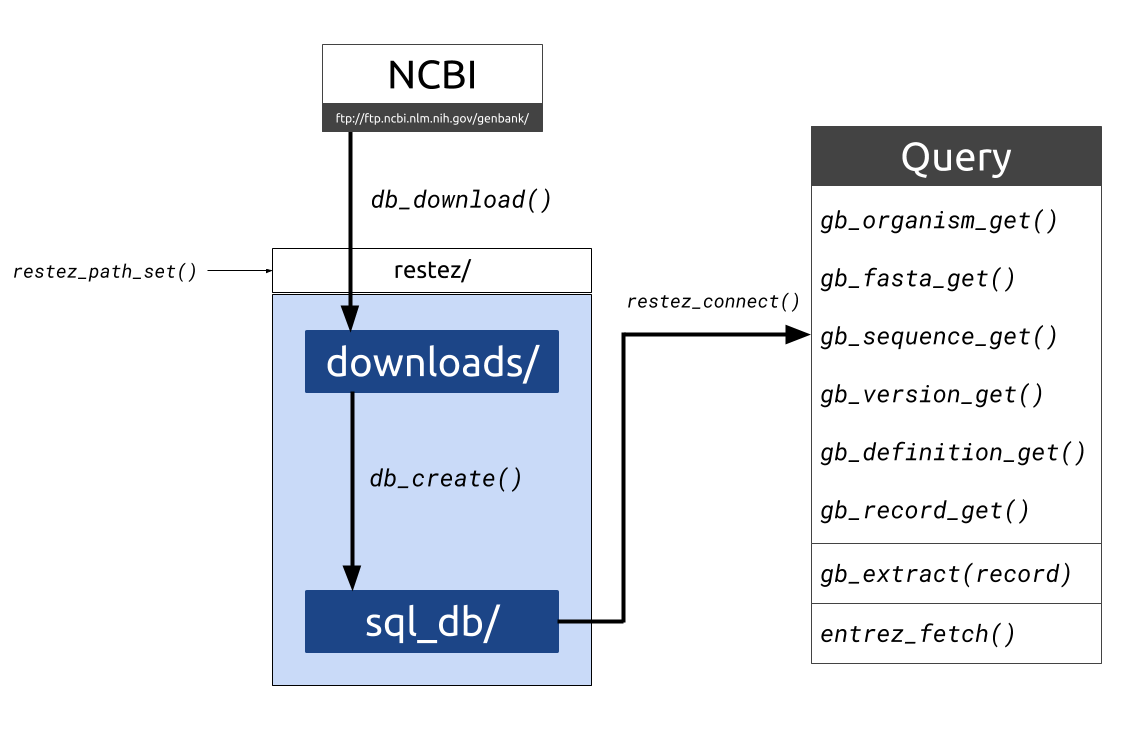
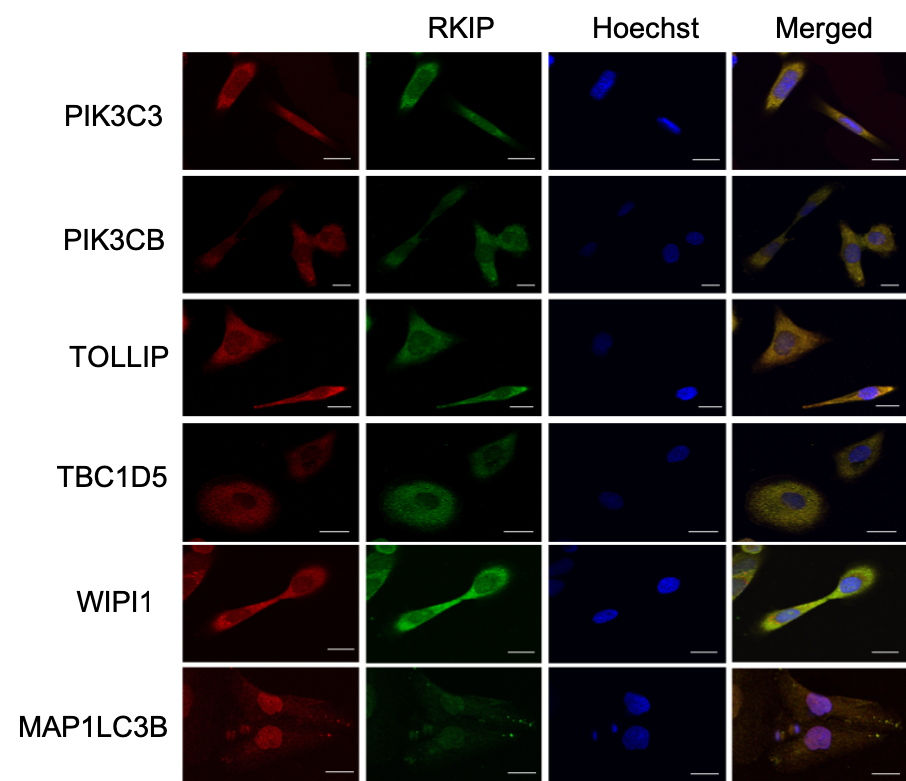
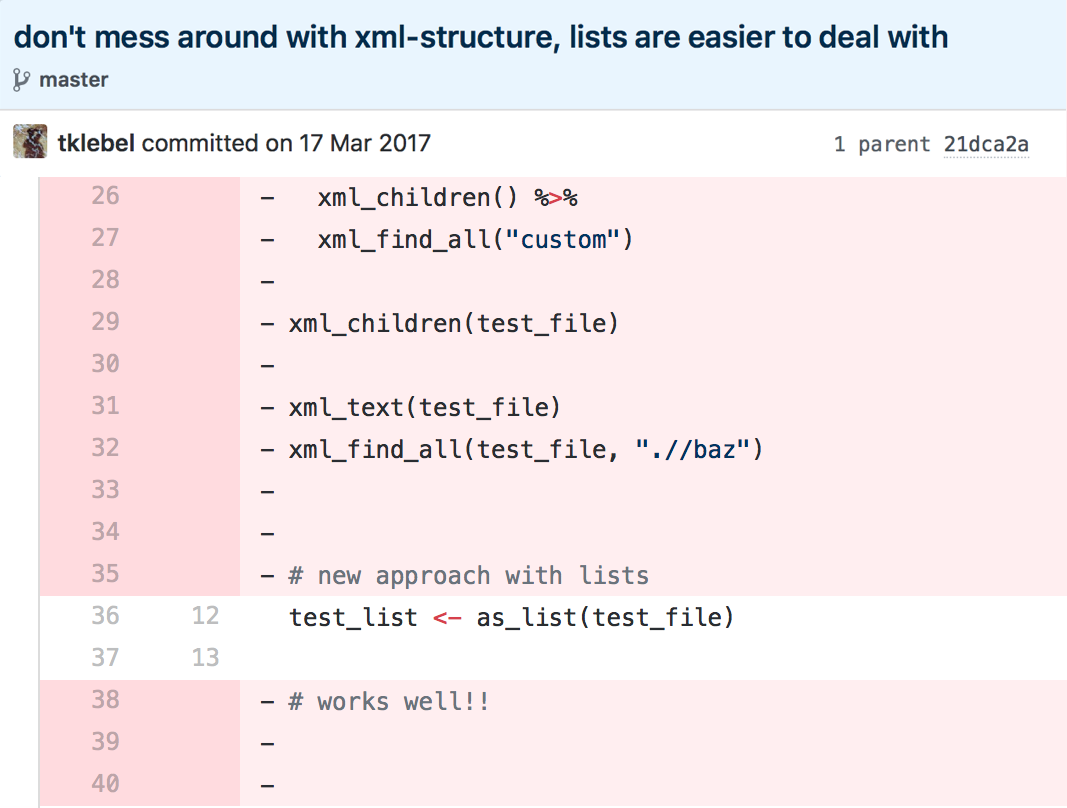
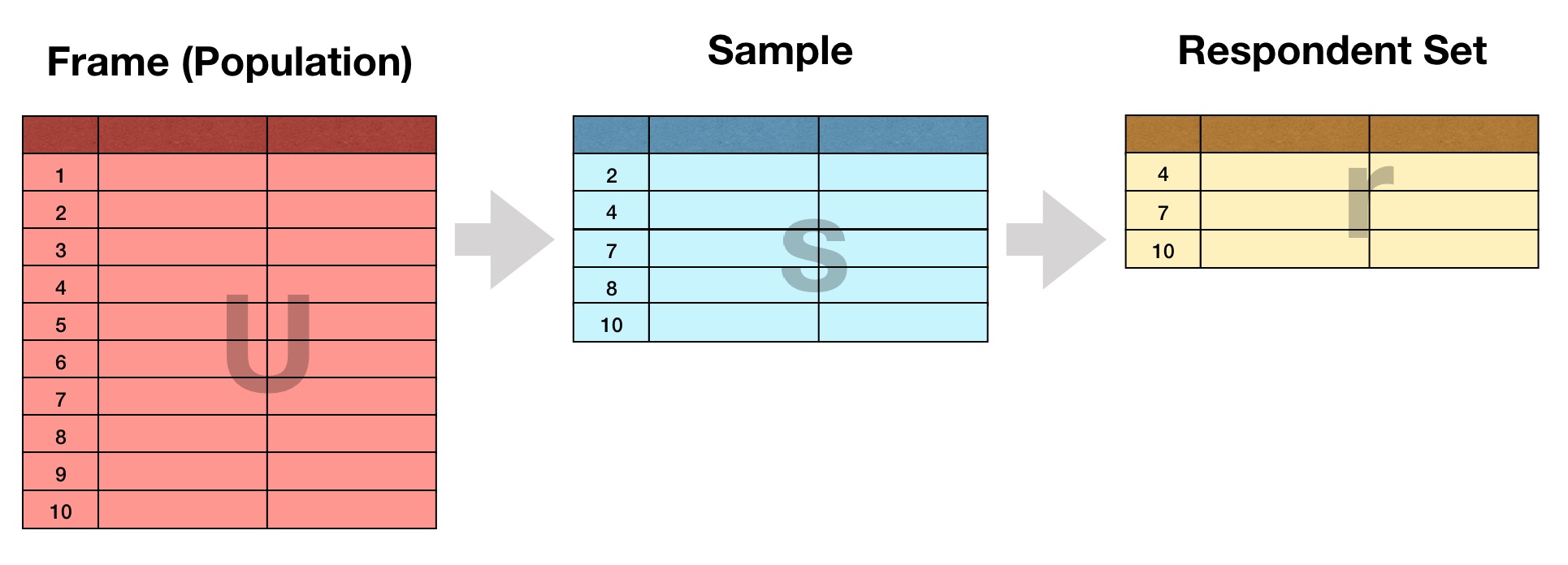
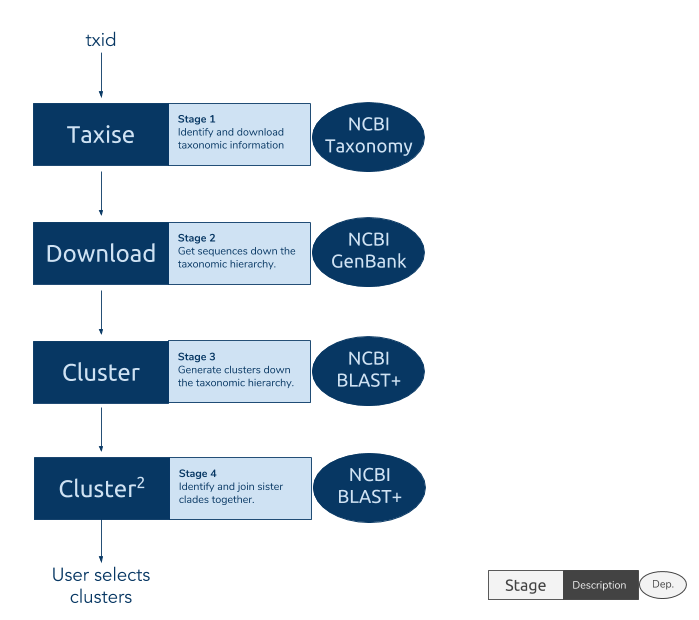
spatsoc is an R package written by Alec Robitaille, Quinn Webber and Eric Vander Wal of the Wildlife Evolutionary Ecology Lab (WEEL) at Memorial University of Newfoundland. It is the lab’s first R package and was recently accepted through the rOpenSci onboarding process with a big thanks to reviewers Priscilla Minotti and Filipe Teixeira, and editor Lincoln Mullen.