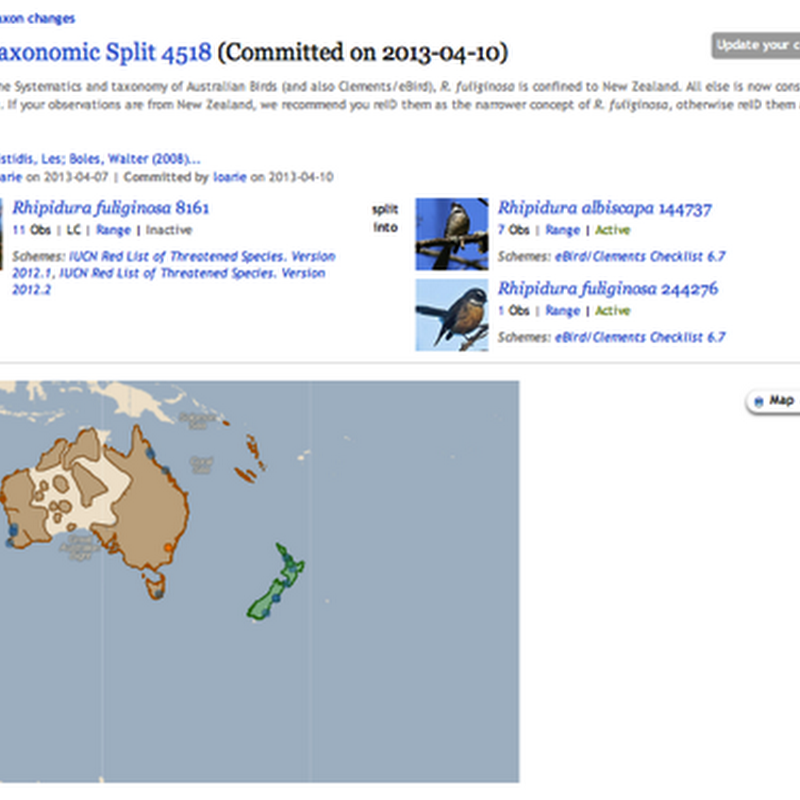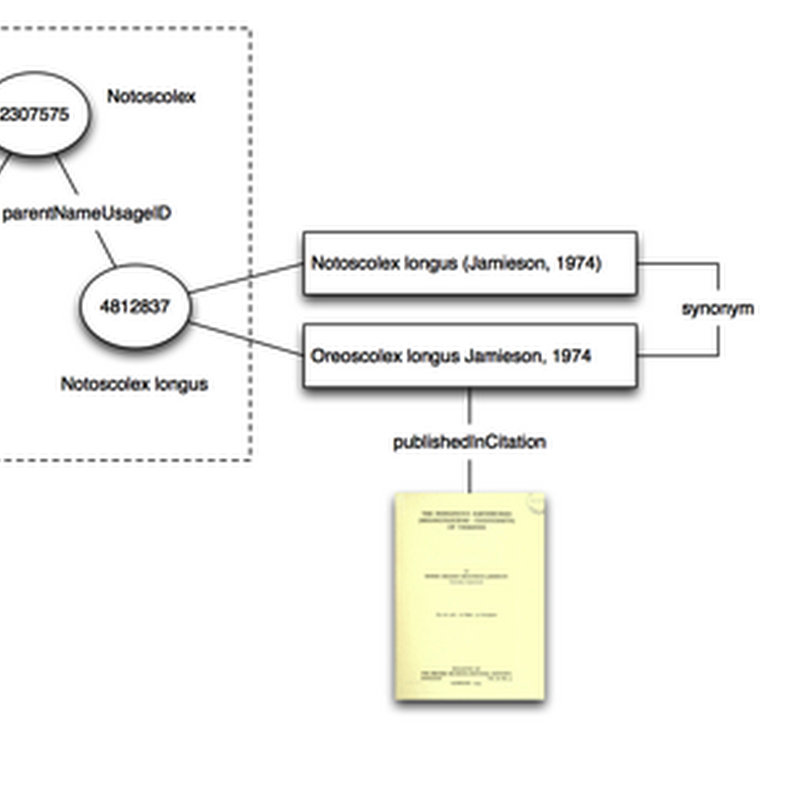
Quick notes on "taxon concepts". In order to navigate through taxon names I plan to have at least one taxonomic classification in BioNames. GBIF makes the most sense at this stage. The model I'm adopting is that the classification is a graph where nodes have the id used by the external database (in this case GBIF). Each node has one or more names attached, and where possible the names are linked to the original description.