Last night I upgraded the software behind Chemical blogspace, to the version online on Google Code, though I needed the help from Eaun to get paper titles correctly picked up for ACS journals.

Last night I upgraded the software behind Chemical blogspace, to the version online on Google Code, though I needed the help from Eaun to get paper titles correctly picked up for ACS journals.
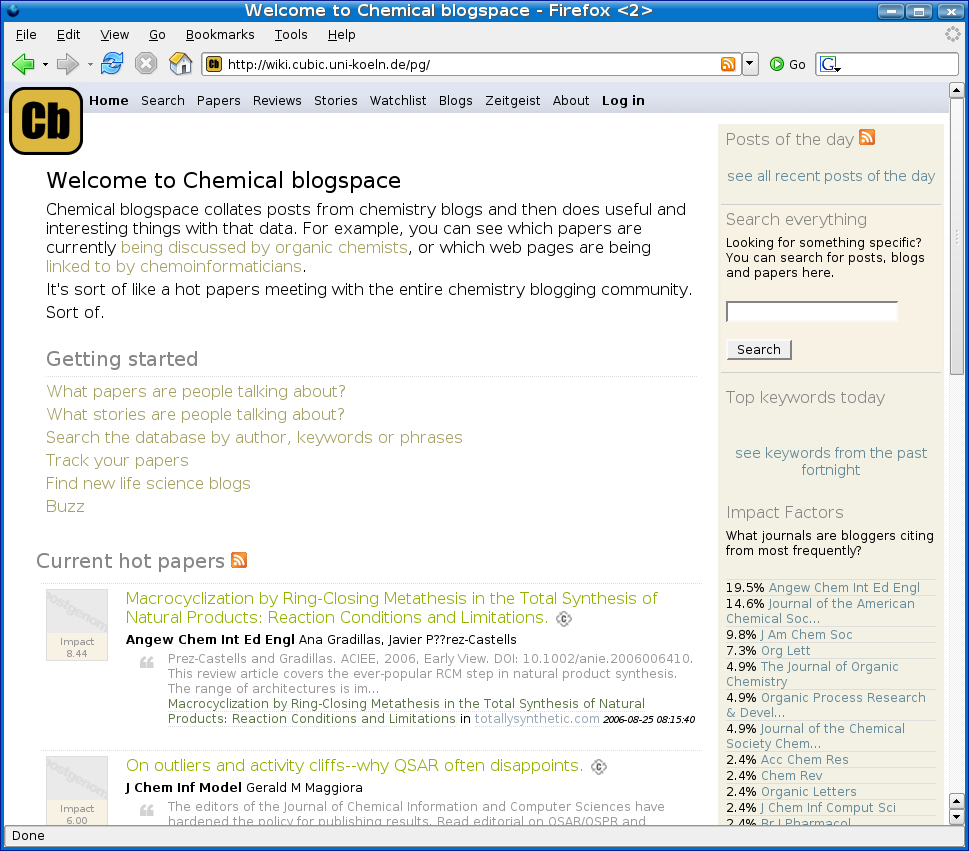
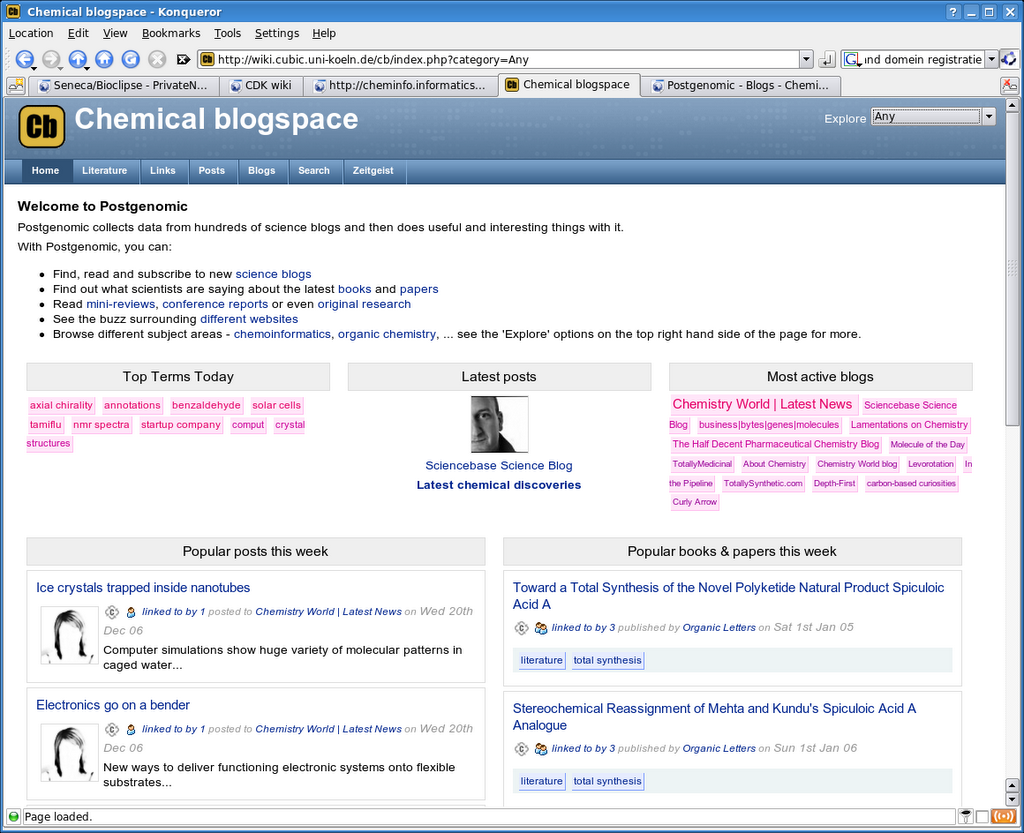
We all know chemical space; Chemical blogspace (Cb) is different: it is the chemistry discussed in blogspace. Cb is build on the opensource software of Postgenomic.com which I bloged on before. The now running Cb aggregates 19 blogs and, like the original, extracts linked (cited or reviewed) articles from literature. The system is beta, but I am happy about it already that I mention it now.
Earlier I reported about postgenomic.com , and needed some diversion from my manuscript work (could no longer think straight about the article I’m working on). So time for some reading up on new technologies. Timing was perfect, because the source code of postgenomic.com got just uploaded to SourceForge SVN.
Roland Krause discussed today in his blog Notes from the Biomass an interesting website: postgenomic.com . This website, still marked BETA, mines blogs in the field of genomics and extract noteworthy statistics from it: which articles are cited in those blogs. For example, the most discussed article is Kai Wang’s Gene-function wiki would let biologists pool worldwide resources in Nature.