I noted earlier this week that [d]uring the week [in Oxford ], someone (name and address is know at the editorial office) commented on the fact that my blog posts are somewhat difficult to follow;
Messaggi di Rogue Scholar
There was some talk about the history of chemoinformatics toolkits by Noel and Andrew, which made me wonder on the exact history of Jmol and JChemPaint.
I hacked in a new extension point for Bioclipse yesterday, based on a proposal I made earlier. The new extension point (EP) is called ChildResourceCreator and allows creating child resources for a given IBioResource. One application where this is very useful is the CMLRSS application (earlier blog), or any RSS or Atom enriched with any other XML language.
Recently, a new generation of Chemical Markup Language CML users seem to hit the learning-curve-wall; there seems to be a niche in explaining the use of CML, so here goes. My new (third) blog will discuss frequently and less frequently asked questions about the use of CML.
Mix one of the oldest and one of the latest computer technologies, and you get FoX (BSD license), a Fortran library for reading and writing Chemical Markup Language, and thus XML.
David Strumfels posted news about the Useful Chemistry CMLRSS feed . He explains how this feed can be accessed using Jmol and Bioclipse. The latter are accompanied by two AVI movies: one about creating a new OPML file, and one about accessing the CMLRSS file from the OPML.
Yesterday I installed the Eclipse Web Tools Platform again, and now succesfully, using the Eclipse update mechanism, on my Kubuntu dapper eclipse install. Because it has a validating XML editor, the one last thing I still needed jEdit for. (I do miss the vertical selection feature of jEdit, though.) It signals me of errors, and allows autocompletion.
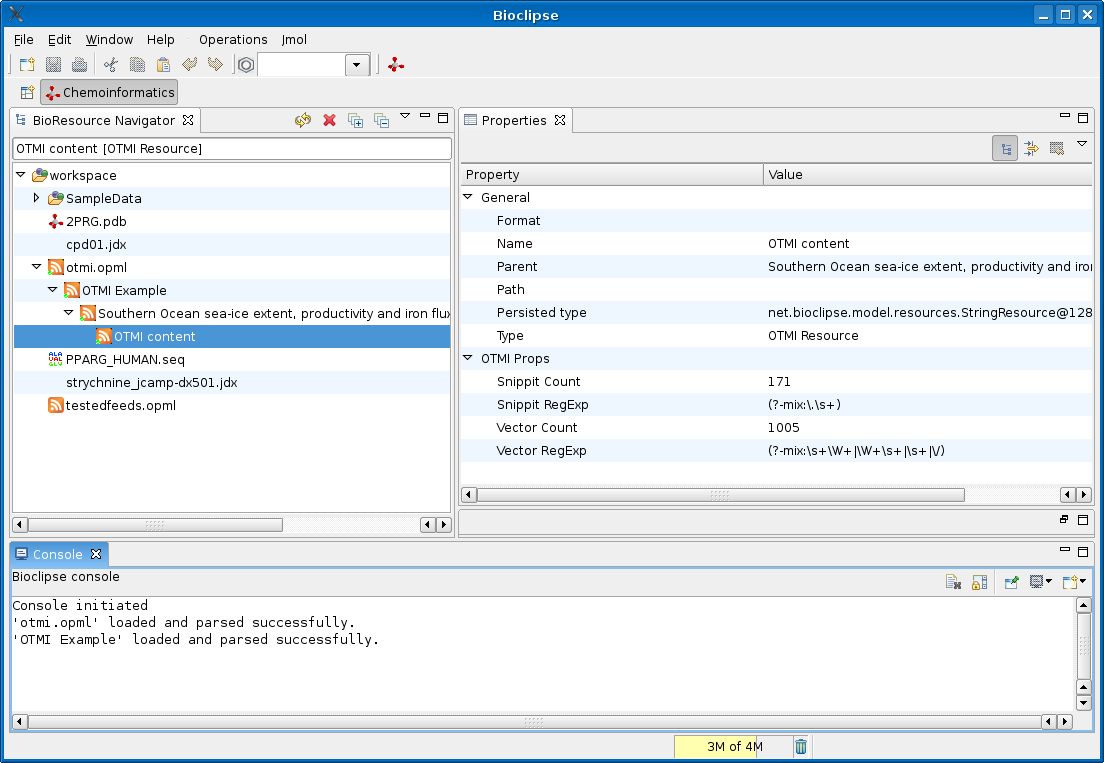
Timo Hannay blogged in Nature’s Nascent blog about the Open Text Mining Interface (OTMI), which is “a suggestion from Nature about how we might achieve text-mining and indexing purposes”. The idea is that each article has a link pointing to a machine readable file containing raw data about (and from?) the article.
As of April 3, I will be working as postdoc in the group of Christoph Steinbeck at the Cologne University BioInformatics Center, or simply CUBIC, for a year. Though no exact plans have been decided upon, the work will include CDK, CML, ontologies, Bioclipse, semantic web technologies, Jmol, and other interesting things. Research areas will at least include QSAR, but I hope to touch bits of bioinformatics too.
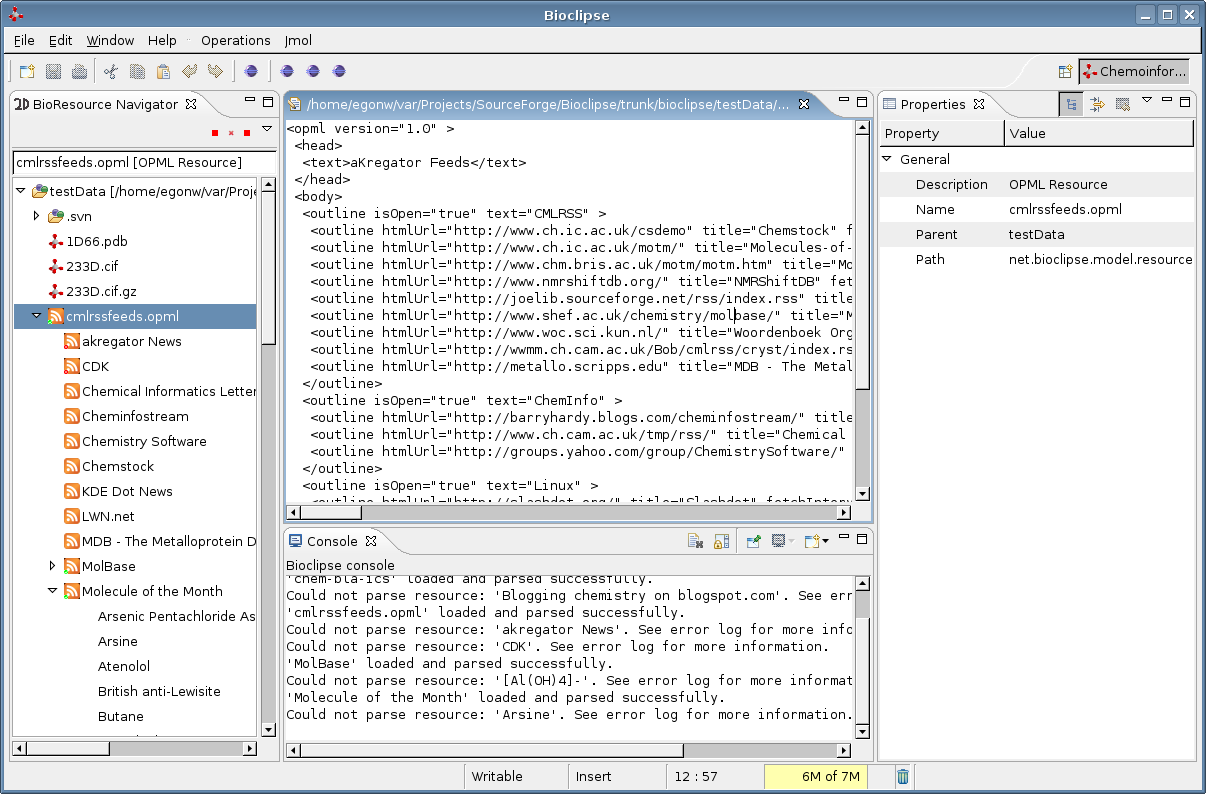
With quite some help from Ola (thanx!), I made good progress with the CMLRSS plugin . The current result looks like: A problem in the transition from Jumbo 5.0 to 5.1 is causing a problem so that it does not show a 3D model or 2D diagram, but that will follow soon.