Quick notes to self following on from a conversation about linking taxonomic names to the literature. There are different sorts of citation: Paper cites another paper Paper cites a dataset Dataset cites a paper Citation type (1) is largely a solved problem (although there are issues of the ownership and use of this data, see e.g. Zootaxa has no impact factor.
Messaggi di Rogue Scholar
In recent years, findable data has become ever more important (the F in FAIR). Here I test that F using the DataCite search service. Firstly an introduction to this service. This is a metadata database about datasets and other research objects.
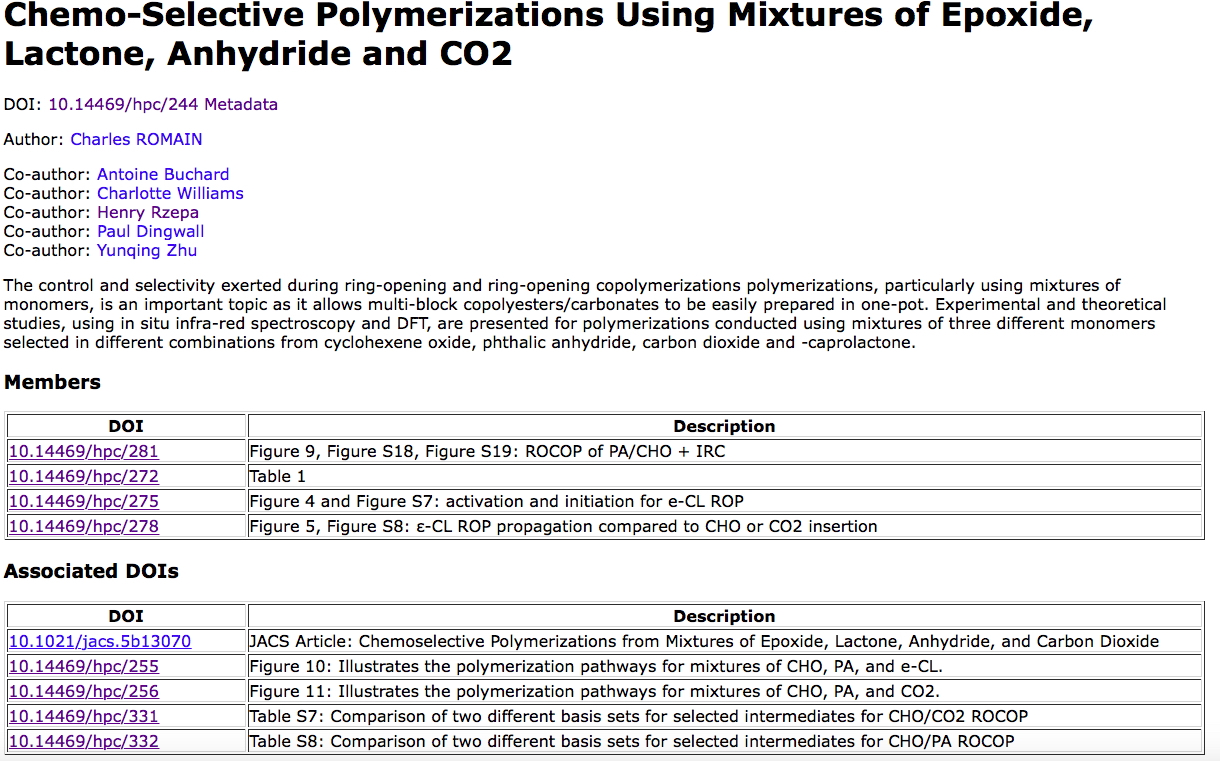
I want to describe a recent attempt by a group of collaborators to share the research data associated with their just published article.[cite]10.1021/jacs.5b13070[/cite] I am here introducing things in a hierarchical form (i.e. not necessarily the serial order in which actions were taken). The data repository selected for the data sharing is described by (m3data) doi: 10.17616/R3K64N[cite]10.17616/R3K64N[/cite] A collaborative

On Friday I discovered that BHL has started issuing CrossRef DOIs for articles, starting with the journal Revue Suisse de Zoologie . The metadata for these articles comes from BioStor. After a WTF and WWIC moment, I tweeted about this, and something of a Twitter storm (and email storm) ensued: To be clear, I'm very happy that BHL is finally assigning article-level DOIs, and that it is doing this via CrossRef.
I had a long Twitter conversation with Terry Catapano (@catapanoth) today, and as can happen with a distracted stream of tweets, I think we were a little at cross purposes. This blog post is an attempt to unpack the debate.

This is a quick sketch of a way to combine existing tools to help clean and annotate data in GBIF, particularly (but not exclusively) occurrence data. GitHub The data provider puts a Darwin Core Archive (expanded, not zipped) into a GitHub repository. GBIF forks the repository, cleans the data, and uploads that to GBIF to populate the database behind the portal.

I've been banging on about having citable, persistent identifiers for specimens, so was suitably impressed when Derek Sikes posted a comment on iPhylo that Arctos already does this. For example, here is a DOI for a specimen: http://dx.doi.org/10.7299/X7VQ32SJ. So, we're all done, right? Not quite.

Regular reader of this blog will be aware of our efforts to promote data citation using digital object identifiers (DOIs), and this week, alongside Rebecca Lawrence from F1000 Research and Kevin Ashley from the Digital Curation Centre, our Editor in Chief Laurie Goodman has a correspondence in Nature strongly making this case.
Readers of this blog will be well versed on our and others work using DataCite Digital Object Identifiers (DOIs) to cite data, and this months DataCite summer meeting in Copenhagen was a good opportunity to take stock of the many recent developments in the area of data publication, with the last six months being particularly busy with the number of new data platforms and data journals announced.

A correspondence we have contributed to has just been published in the BMC Research Notes “Data standardization, sharing and publication series” on the adventures in data-citation and data-release practices surrounding the Sorghum genome that is available in our GigaDB database and that was published last year in Genome Biology . We use Sorghum as an example to highlight the issues surrounding data release and use strong words,