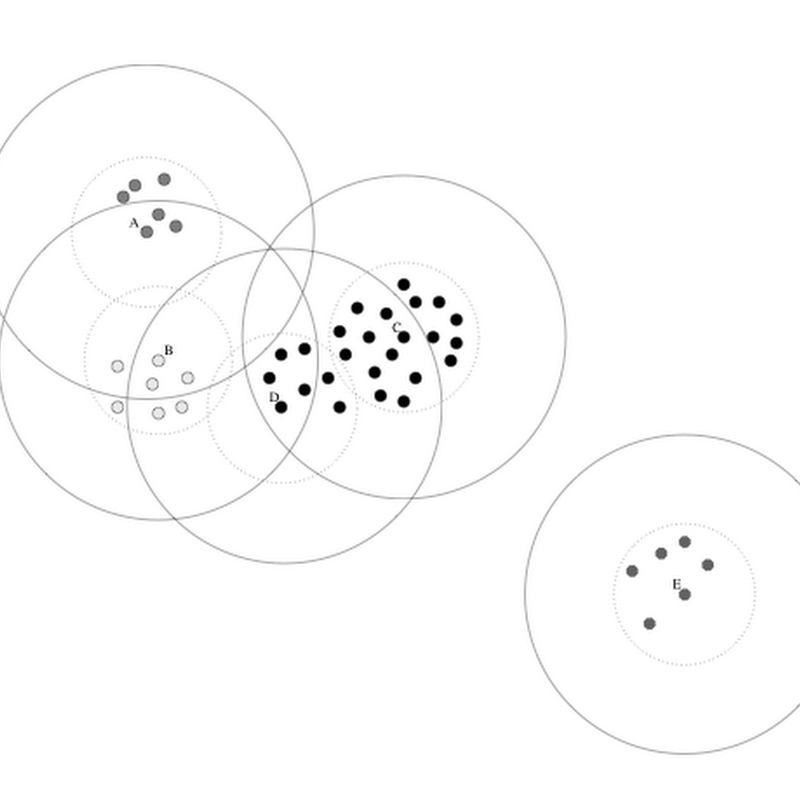
Over on Google Plus (yeah, me neither) Donat Agosti is giving me a hard time regarding the quality of some data that I am using. I've responded to Donat directly, but here I just want to quickly outline two different approaches to cleaning and reconciling bibliographic metadata.