Note to self (basically rewriting last year's Finding citations of specimens). Bibliographic data supports going from identifier to citation string and back again, so we can do a "round trip." 1. Given a DOI we can get structured data with a simple HTTP fetch, then use a tool such as citation.js to convert that data into a human-readable string in a variety of formats.
Messaggi di Rogue Scholar

I've put a short note up on bioRxiv about ways to geocode nucleotide sequences in databases such as GenBank. The preprint is "Geocoding genomic databases using GBIF" https://doi.org/10.1101/469650.

This post is a response to Ross Mounce's post Text mining for museum specimen identifiers. As Ross notes in that post, mining literature for specimen codes is something I've been interested in for a while (search for specimen codes on iPhylo), and @Aime Rankin (formerly an undergraduate student at Glasgow) did some work on this as well. It's great to see progress in this area.

Playing with the my "material examined" tool I've been working on, I wondered whether I could make use of it in, say, a spreadsheet. Imagine that I have a spreadsheet of museum codes and want to look those up in GBIF. I could create a service for Open Refine but Open Refine is a bit big and clunky, you have to fire up a Java application and point your browser at it, and Open Refine isn't as intuitive or as flexible as a spreadsheet.
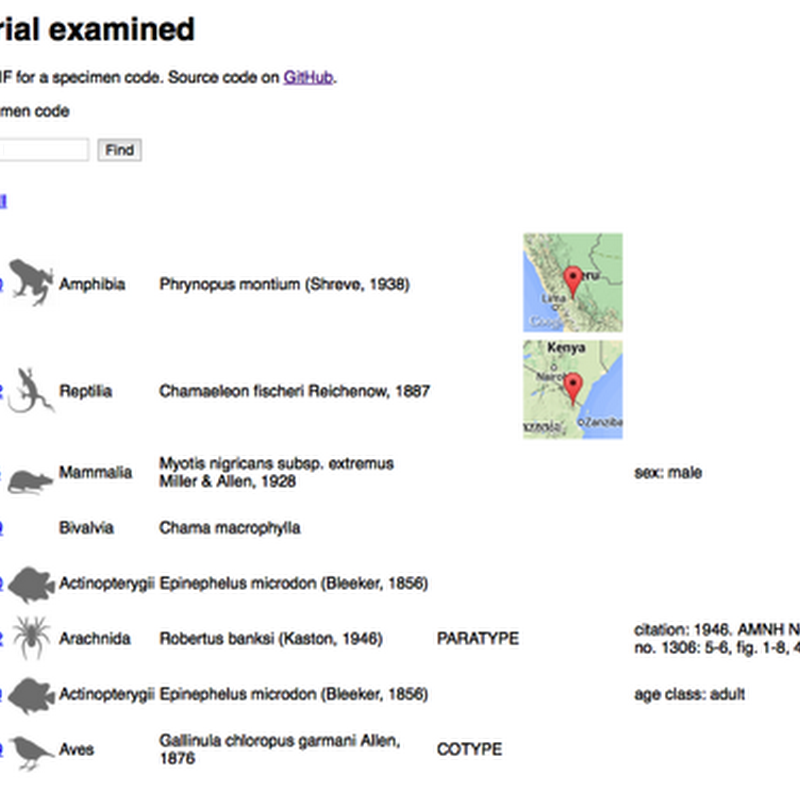
I've put together a working demo of some code I've been working on to discover GBIF records that correspond to museum specimen codes. The live demo is at http://bionames.org/~rpage/material-examined/ and code is on GitHub. To use the demo, simply paste in a specimen code (e.g., "MCZ 24351") and click Find and it will do it's best to parse the code, then go off to GBIF and see what it can find.

One reason I'm excited by the launch of the NHM data portal is that it opens up opportunities to link publications about specimens i the NHM to the record of the specimens themselves.
Based on recent discussions my sense is that our community will continue to thrash the issue of identifiers to death, repeating many of the debates that have gone on (and will go on) in other areas. To be trite, it seems to me we have three criteria: cheap , resolvable , and persistent . We get to pick two.

Duplicate records are the bane of any project that aggregates data from multiple sources.
Quick note about a tool I've cobbled together as part of the phyloinformatics course, which addresses a long standing need I and others have to extract specimen codes from text. I've had this code kicking around for a while (as part of various never-finished data mining projects), but never got around to releasing it, until now.