So it looks like TreeBASE is in trouble, it's legacy Java code a victim of security issues. Perhaps this is a chance to rethink TreeBASE, assuming that a repository of published phylogenies is still considered a worthwhile thing to have (and I think that question is open). Here's what I think could be done.
Rogue Scholar Posts
babette 1 is a package to work with BEAST2 2 ,a software platform for Bayesian evolutionary analysis from R. babette is a spin-off of my own academic research.As a PhD I work on models of diversification: mathematical descriptionsof how species form new species.
Phylogenetic trees are commonly used to present evolutionary relationships of species. Newick is the de facto format in phylogenetic for representing tree(s). Nexus format incorporates Newick tree text with related information organized into separated units known as blocks. For the R community, we have ape and phylobase packages to import trees from Newick and Nexus formats.
Notes to self on web map-style tree viewers. The basic idea is to use Google Maps or Leaflet to display a tree. Hence we need to compute tiles. One approach is to use a database that supports spatial queries to store the x,y coordinates of the tree. When we draw a tile we compute the coordinates of that tile, based on position and zoom level, do a spatial query to extract all lines that intersect with the rectangle for that tile, and draw those.

Some notes to self about future directions for the "million DNA barcodes map" http://iphylo.org/~rpage/bold-map/. At the moment we have an interactive map that we can pan and zoom, and click on a marker to get a list of one or more barcodes at the location. We can also filter by major taxonomic group. Here are some ideas on what could be next. Search At the moment search is simply browsing the map.
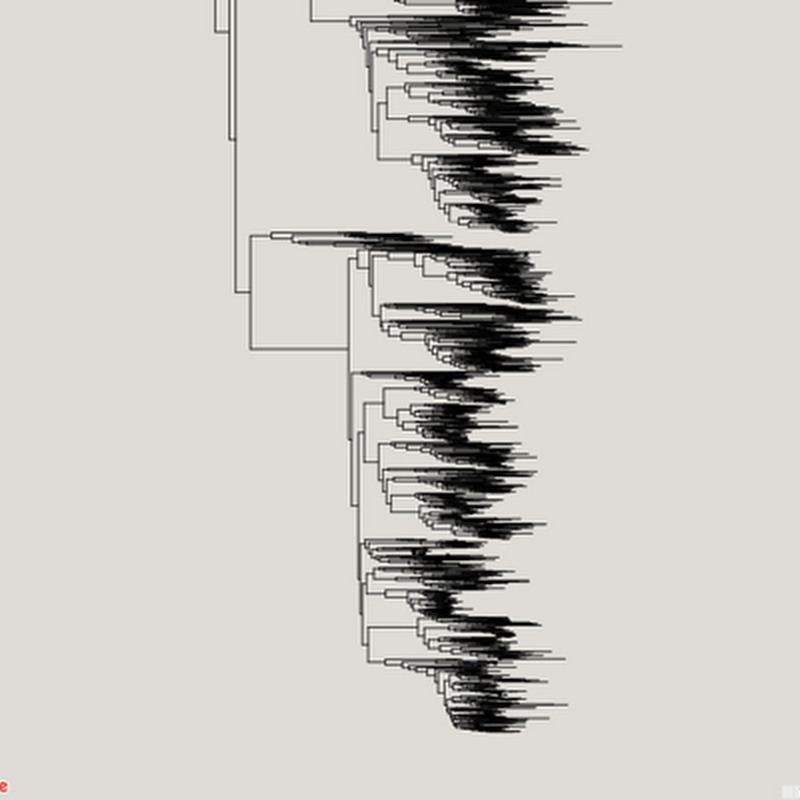
Inspired in part by the release of the draft tree of life (doi:10.1073/pnas.1423041112 by the Open Tree of Life, I've been revisiting (yet again) ways to visualise very big phylogenies (see Very large phylogeny viewer for my last attempt). My latest experiment uses Google Maps to render a large tree. Google Maps uses "tiles" to create a zoomable interface, so we need to create tiles for different zoom levels for the phylogeny.

Currently in classes where I teach the basics of tree building, we still fire up ancient iMacs, load up MacClade, and let the students have a play. Typically we give them the same data set and have a class competition to see which group can get the shortest tree by manually rearranging the branches. It’s fun, but the computers are old, and what’s nostalgic for me seems alien to the iPhone generation.
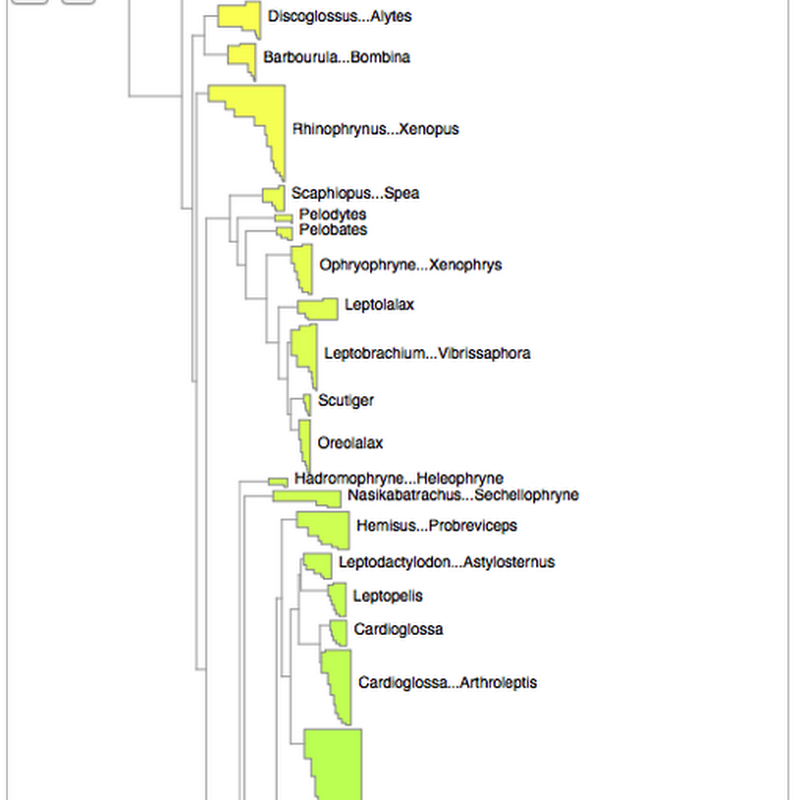
As announced on phylobabble I've started to revisit visualising large phylogenies, building on some work I did a couple of years ago (my how time flies). This time, there is actual code (see https://github.com/rdmpage/deep-tree) as well as a live demo http://iphylo.org/~rpage/deep-tree/demo/. You can see the amphibian tree below at http://iphylo.org/~rpage/deep-tree/demo/show.php?id=5369171e32b7a:You can upload or paste a tree (for now in NEXUS
Bryan Drew and colleagues have published a piece in PLoS Biology bemoaning the lack of databased phylogenies: This is an old problem (see for example "Towards a Taxonomically Intelligent Phylogenetic Database" doi:10.1038/npre.2007.1028.1), but alas the solution proposed by Drew et al. is also old:In my opinion, as soon as you start demanding people do something you've lost the argument, and you're relying on power ("you don't get to publish

My latest tweak to BioNames is to add colour to the phylogenies. Terminal nodes with the same name are labelled with the same background colour. For example, here is a tree for fiddler and ghost crabs:The colours make it easier to see that this tree has a mixture of a few sequences from divergent taxa, and a lot of sequences from the same taxa.Note that you can now also download the SVG drawing of the tree.