phylogram: dendrograms for evolutionary analysis
Creators
Evolutionary biologists are increasingly using R for building,editing and visualizing phylogenetic trees.The reproducible code-based workflow and comprehensive array of toolsavailable in packages such as ape,phangorn andphytools make R an ideal platform forphylogenetic analysis.Yet the many different tree formats are not well integrated,as pointed out in a recentpost.
The standard data structure for phylogenies in R is the "phylo"object, a memory efficient, matrix-based tree representation.However, non-biologists have tended to use a tree structurecalled the "dendrogram", which is a deeply nested list withnode properties defined by various attributes stored at each level.While certainly not as memory efficient as the matrix-based format,dendrograms are versatile and intuitive to manipulate, and hencea large number of analytical and visualization functions existfor this object type. A good example is thedendextend package,which features an impressive range of options for editing dendrogramsand plotting publication-quality trees.
To better integrate the phylo and dendrogram object types,and hence increase the options available for both camps,we developed the phylogrampackage, which is now a part of the rOpenSciproject.This small package features a handful of functions for tree conversion,importing and exporting trees as parenthetic text, and manipulatingdendrograms for phylogenetic applications.The phylogram package draws heavily on ape,but currently has no other non-standard dependencies.
\
🔗Installation
To download phylogram from CRAN and load the package, run
install.packages("phylogram")library(phylogram)Alternatively, to download the latest development version from GitHub,first ensure that the devtools,kmer, anddendextendpackages are installed,then run:
devtools::install_github("ropensci/phylogram", build_vignettes = TRUE) library(phylogram)\
🔗Tree import/export
A wide variety of tree formats can be parsed as phylo objects using either thewell-optimized ape::read.tree function(for Newickstrings),or the suite of specialized functions in the versatiletreeio package.To convert a phylo object to a dendrogram, the phylogram package includesthe function as.dendrogram, which retains node height attributes and can handlenon-ultrametric trees.
For single-line parsing of dendrograms from Newick text,the read.dendrogram function wraps ape::read.treeand converts the resulting phylo class object to a dendrogram using as.dendrogram.
Similarly, the functions write.dendrogram and as.phylo are used toexport dendrogram objects to parenthetic text and phylo objects, respectively.
\
🔗Tree editing
The phylogram package includes some new functions for manipulatingtrees in dendrogram format.Leaf nodes and internal branching nodes can be removedusing the function prune, which identifies andrecursively deletes nodes based on patternmatching of "label" attributes.This is slower than ape::drop.tip, but offersthe benefits of versatile string matching using regular expressions,and the ability to remove inner nodes (and by extension all subnodes)that feature matching "label" attributes.To aid visualization, the function ladder rearrangesthe tree, sorting nodes by the number of members(analogous to ape::ladderize).
For more controlled subsetting or when creating trees from scratch(e.g. from a standard nested list), the function remidpointrecursively corrects all "midpoint", "members" and "leaf" attributes.Node heights can then be manipulated using either reposition, whichscales the heights of all nodes in a tree by a given constant, oras.cladogram, which resets the "height" attributes of all terminalleaf nodes to zero and progressively resets the heights of the inner nodesin single incremental units.
As an example, a simple three-leaf dendrogram can be created froma nested list as follows:
x <- list(1, list(2, 3))## set class, midpoint, members and leaf attributes for each nodex <- remidpoint(x)## set height attributes for each nodex <- as.cladogram(x)A nice feature of the dendrogram object type is that treeediting operations can be carried out recursivelyusing fast inbuilt functions in the "apply" family such as dendrapplyand lapply.
For example, to label each leaf node of the tree alphabetically we cancreate a simple labeling function and apply it to the tree nodes recursively usingdendrapply.
set_label <- function(node){ if(is.leaf(node)) attr(node, "label") <- LETTERS[node] return(node)}x <- dendrapply(x, set_label)plot(x, horiz = TRUE)
🔗Applications
One application motivating bi-directional conversion between phylo anddendrogram objects involves creating publication-quality 'tanglegrams' usingthe dendextend package.For example, to see how well the fast, alignment-free k-mer distancefrom the kmer packageperforms in comparison to the standard Kimura 1980 distance measure,we can create neighbor-joining trees using each method and plot them side by sideto check for incongruent nodes.
## load woodmouse data and remove columns with ambiguitiesdata(woodmouse, package = "ape")woodmouse <- woodmouse[, apply(woodmouse, 2, function(v) !any(v == 0xf0))]## compute Kimura 1980 pairwise distance matrixdist1 <- ape::dist.dna(woodmouse, model = "K80")## deconstruct alignment (not strictly necessary)woodmouse <- as.list(as.data.frame(unclass(t(woodmouse))))## compute kmer distance matrix dist2 <- kmer::kdistance(woodmouse, k = 7) ## build and ladderize neighbor-joining treesphy1 <- ape::nj(dist1)phy2 <- ape::nj(dist2)phy1 <- ape::ladderize(phy1)phy2 <- ape::ladderize(phy2)## convert phylo objects to dendrogramsdnd1 <- as.dendrogram(phy1)dnd2 <- as.dendrogram(phy2)## plot the tanglegramdndlist <- dendextend::dendlist(dnd1, dnd2)dendextend::tanglegram(dndlist, fast = TRUE, margin_inner = 5)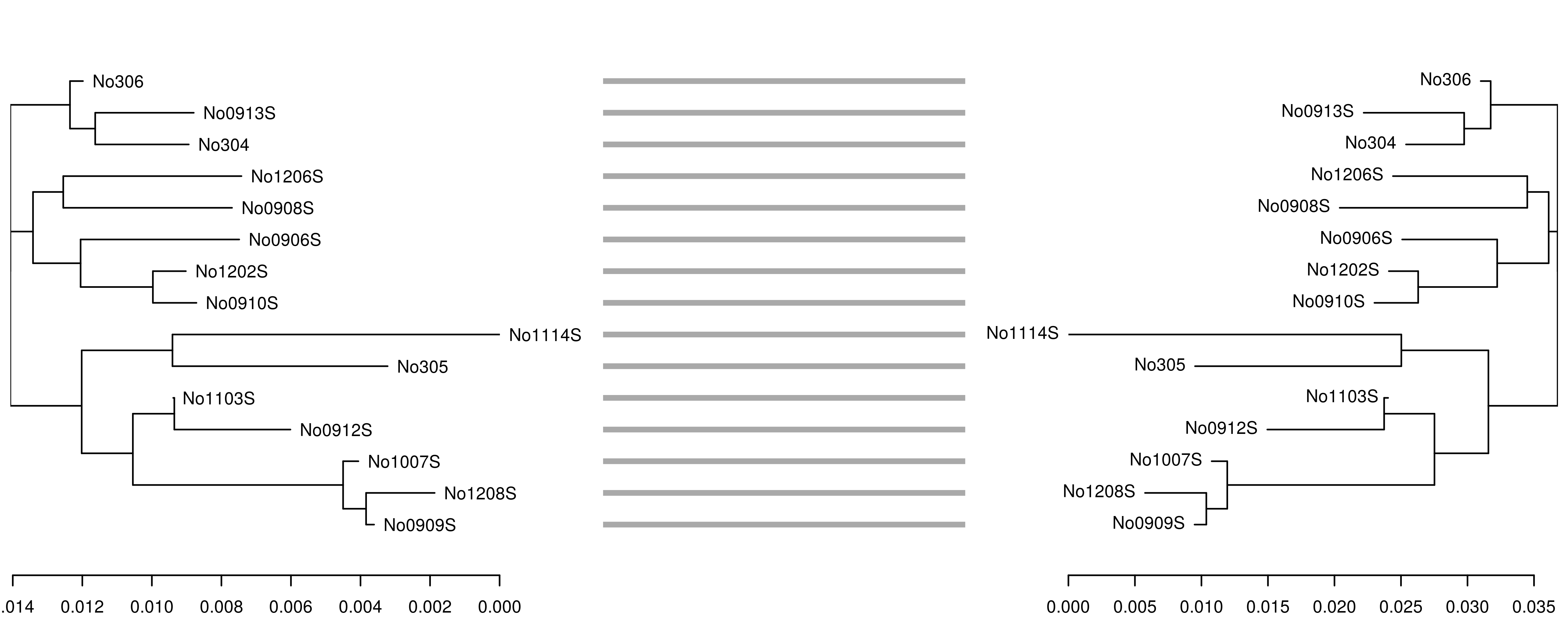
\
In this case, the trees are congruent and branch lengths are similar.However, if we reduce the k-mer size from 7 to 6,the accuracy of the tree reconstruction is affected, as shown by theincongruence between the original K80 tree (left) and the tree derivedfrom the 6-mer distance matrix (right):
## compute kmer distance matrix dist3 <- kmer::kdistance(woodmouse, k = 6) phy3 <- ape::nj(dist3)phy3 <- ape::ladderize(phy3)dnd3 <- as.dendrogram(phy3)dndlist <- dendextend::dendlist(dnd1, dnd3)dendextend::tanglegram(dndlist, fast = TRUE, margin_inner = 5)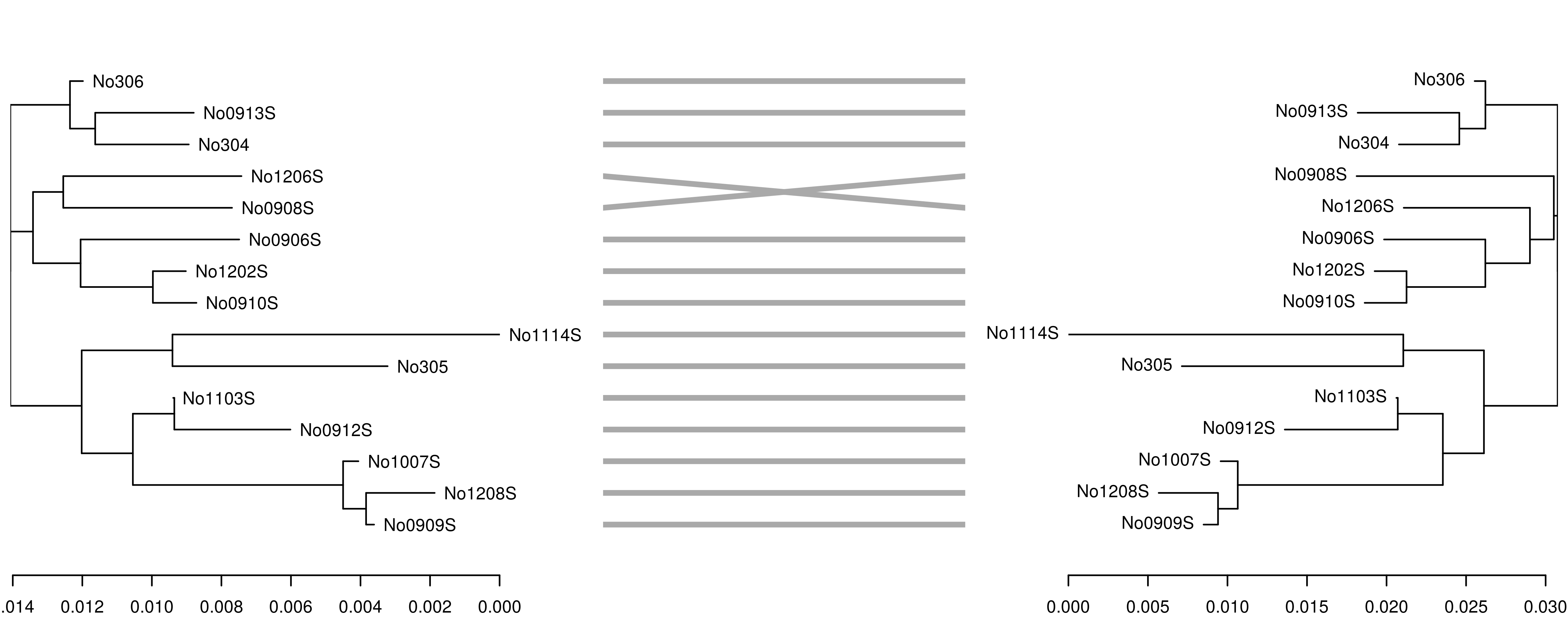
\
Hopefully users will find the package useful for a range of other applications.Bug reports and other suggestions are welcomed, and can be directed to theGitHub issues pageor the phylogram google group.Thanks to Will Cornwell andBen J. Wardfor reviewing the code and suggesting improvements,and to Scott Chamberlainfor handling the rOpenScionboarding process.
The phylogram package is available for download fromGitHub andCRAN,and a summary of the package is published in theJournal of Open Source Software.
Additional details
Description
Evolutionary biologists are increasingly using R for building,editing and visualizing phylogenetic trees.The reproducible code-based workflow and comprehensive array of toolsavailable in packages such as ape,phangorn andphytools make R an ideal platform forphylogenetic analysis.Yet the many different tree formats are not well integrated,as pointed out in a recentpost.
Identifiers
- UUID
- e000a191-7701-4481-a139-b1e0fd8179d2
- GUID
- https://doi.org/10.59350/rg4rw-jjv50
- URL
- https://ropensci.org/blog/2018/07/12/phylogram/
Dates
- Issued
-
2018-07-12T00:00:00Z
- Updated
-
2025-02-13T12:42:17Z