A recurrent topic among faculty and librarians interested in infrastructure reform is the question of whose turn it is to make the next move. Researchers rightfully argue that they cannot submit their work exclusively to modern, open access journals because that would risk their and their employees’ jobs. Librarians, equally correctly, argue that they would cancel subscriptions if faculty wouldn’t complain about ensuing access issues.
Messaggi di Rogue Scholar

Björn Grüning modelling one of our t-shirts. Nowadays, massive amounts of diverse data are generated in biomedical research. To manage it and extract useful information, bioinformatic solutions are needed and software must be developed. The development of a tool should always follow a similar process.

PH.D STUDENT OPENING IN COMMUNITY ECOLOGY IN THE ERNEST LAB {.wp-image-1708 .alignleft loading=“lazy” decoding=“async” attachment-id=“1708” permalink=“https://jabberwocky.weecology.org/2016/09/19/phd-student-opening-community-ecology-ernest-lab/20916401782_019d3c14eb_z/” orig-file=“https://i0.wp.com/jabberwocky.weecology.org/wp-content/uploads/2016/09/20916401782_019d3c14eb_z.jpg?fit=640%2C480&ssl=1” orig-size=“640,480”

**Deep (ocean) sequencing. Big-(fish) data. **The ocean sunfish, must officially be one of the world’s weirdest creatures to enter the “genome club”, and have its genetic code mapped. Laying the most eggs of any other known vertebrate (up to 300,000,000 at a time), and starting out as the size of the head of a pin, sunfish grow to become to largest bony fish in the sea.

Gigascience this week published a high quality genome of the apple, the “golden delicious” variety, to be precise. Although a version of the Malus domestica genome has already been available, Xuewei Li et al .’s de novo assembly significantly improves on that previous ressource, using a healthy injection of long reads from third generation, “single-molecule” sequencing technology.
The next installment from the Portal Blog by my student Joan Meiners (@beecycles) on how we shook things up at the Portal Project so we could study regime shifts.

At various meetings I get often asked by early career researchers, librarians or other colleagues what my interactions with publishers felt like. I usually answer that my last twelve years campaigning for infrastructure reform felt like academia was receiving the big middle finger from publishers: No matter what you try, academia, we’ll still get your money, stupid suckers!

At seemingly every possibility in a discussion on peer-review, people apparently feel the need to emphasize that in the current model reviewers (or most academic editors handling peer-review) are not being paid.
I received an invitation to SciFoo (a “Science F riends o f O ’Reilly” meeting) in the beginning of April this year. I was aware of the hype around this “unconference” and several people I knew had already participated, so at first I was rather excited and flattered about the invitation. However, I was in Germany, the meeting was at Google headquarters in Palo Alto, California and the flight was not covered.
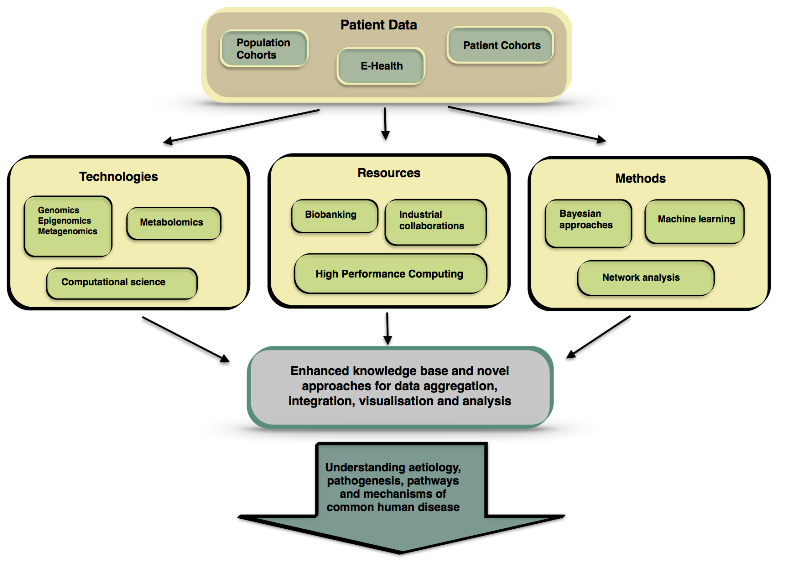
Building an open, community-supported, e-infrastructure for medical metabolomics data. The European Union’s Horizon 2020 research and innovation programme is funding the PhenoMeNal (Phenome and Metabolome aNalysis) project that aims to support data processing and analysis pipelines for molecular phenotype data generated from metabolomics applications.