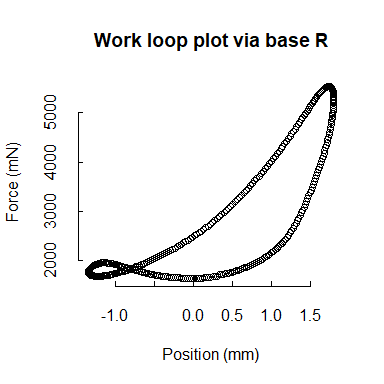
Studies of muscle physiology often rely on closed-source, proprietary software for not only recording data but also for data wrangling and analyses. Although specialized software might be necessary to record data from highly-specialized equipment, data wrangling and analyses should be free from this constraint.