We are pleased to announce the release of our new Code of Conduct. rOpenSci’s community is our best asset and it’s important that we put strong mechanisms in place before we have to act on a report. As before, our Code applies equally to members of the rOpenSci team and to anyone from the community at large participating in in-person or online activities.
Postagens de Rogue Scholar

In late November 2018, we ran the third annual rOpenSci ozunconf. This is the sibling rOpenSci unconference, held in Australia. We ran the first ozunconf in Brisbane in 2016, and the second in Melbourne in 2017. Photos taken by Ajay from Fotoholics As usual, before the unconf, we started discussion on GitHub issue threads,and the excitement was building with the number of issues.

I never really thought I would write an R package. I use R pretty casually. Then, this year, I was invited to participate during the last week of the Analytical Paleobiology short course, an intensive month-long experience in quantitative paleontology. I was thrilled to be invited.

🎤 Dan Sholler, rOpenSci Postdoctoral Fellow 🕘 Tuesday, December 18, 2018, 10-11AM PST; 7-8PM CET (find your timezone) ☎️ Details for joining the Community Call. Everyone is welcome. No RSVP needed. Researchers use open source software for the capabilities it provides, such as streamlined data access and analysis and interoperability with other pieces of the scientific computing ecosystem.
[Scientific communication does not stop at the moment of publication. Scientific discussions in the form of post-publication peer review can provide valuable insights for published articles, bring up an alternative research perspective, or even present updates for published research.

spatsoc is an R package written by Alec Robitaille, Quinn Webber and Eric Vander Wal of the Wildlife Evolutionary Ecology Lab (WEEL) at Memorial University of Newfoundland. It is the lab’s first R package and was recently accepted through the rOpenSci onboarding process with a big thanks to reviewers Priscilla Minotti and Filipe Teixeira, and editor Lincoln Mullen.
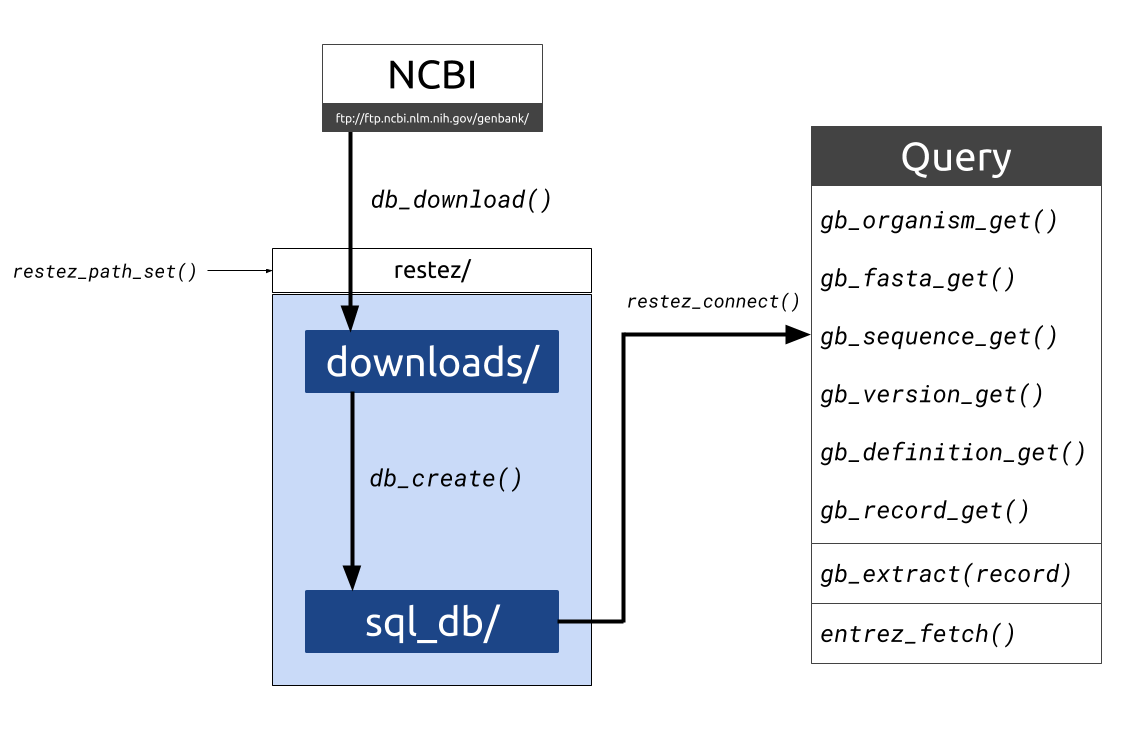
What is restez? R packages for interacting with the National Center for Biotechnology Information (NCBI) have, to-date, depended on API query calls via NCBI’s Entrez.For computational analyses that require the automated look-up of reams of biological sequence data, piecemeal querying via bandwith-limited requests is evidently not ideal.
Although there are increasing incentives and pressures for researchers to share code (even for projects that are not essentially computational), practices vary widely and standards are mostly non-existent. The practice of reviewing code then falls to researchers and research groups before publication.

Antarctic/Southern Ocean science and rOpenSci Collaboration and reproducibility are fundamental to Antarctic and Southern Ocean science, and the value of data to Antarctic science has long been promoted. The Antarctic Treaty (which came into force in 1961) included the provision that scientific observations and results from Antarctica should be openly shared.

While many people groan at the thought of participating in a group ice breaker activity, we’ve gotten consistent feedback from people who have been to recent rOpenSci unconferences. We’ve had lots of requests for a detailed description of how we do it. This post shares our recipe, including a script you can adapt, a reflection on its success, examples of how others have used it, and some tips to remember.