The recently released development draft for SHared Access Research Ecosystem (SHARE), authored by the Association of American Universities (AAU), the Association of Public and Land-grant Universities (APLU) and the Association of Research Libraries (ARL) in response to the OSTP memo on public access to federally funded research in the US sounds a lot like the library-based publishing system I’ve been perpetually arguing for.
Postagens de Rogue Scholar
In our new lab here in Regensburg, we are currently re-establishing the method of confocal microscopy. To start with, we used the fly lines which show a defect in the temporal structure of their spontaneous behavior (a project of one of our graduate students). this fly line uses two transgenic constructs (c105 and c232) to drive expression in specified neurons in the fly brain, i.e., the merged expression patterns of c105 and c232.
I’m a big fan of preprints, the posting of papers in public archives prior to peer review. Preprints speed up the scientific dialogue by letting everyone see research as it happens, not 6 months to 2 years later following the sometimes extensive peer review process. They also allow more extensive pre-publication peer review because input can be solicited from the entire community of scientists, not just two or three individuals.

Insect goo aids biodiversity research Apologies to Jonathan Eisen (see Badomics in the journal), but today in GigaScience we publish a new “squishomics” approach for assessing and understanding biodiversity, using the slightly wacky sounding method of combining DNA-soup made from crushed-up insects and the latest sequencing technology.
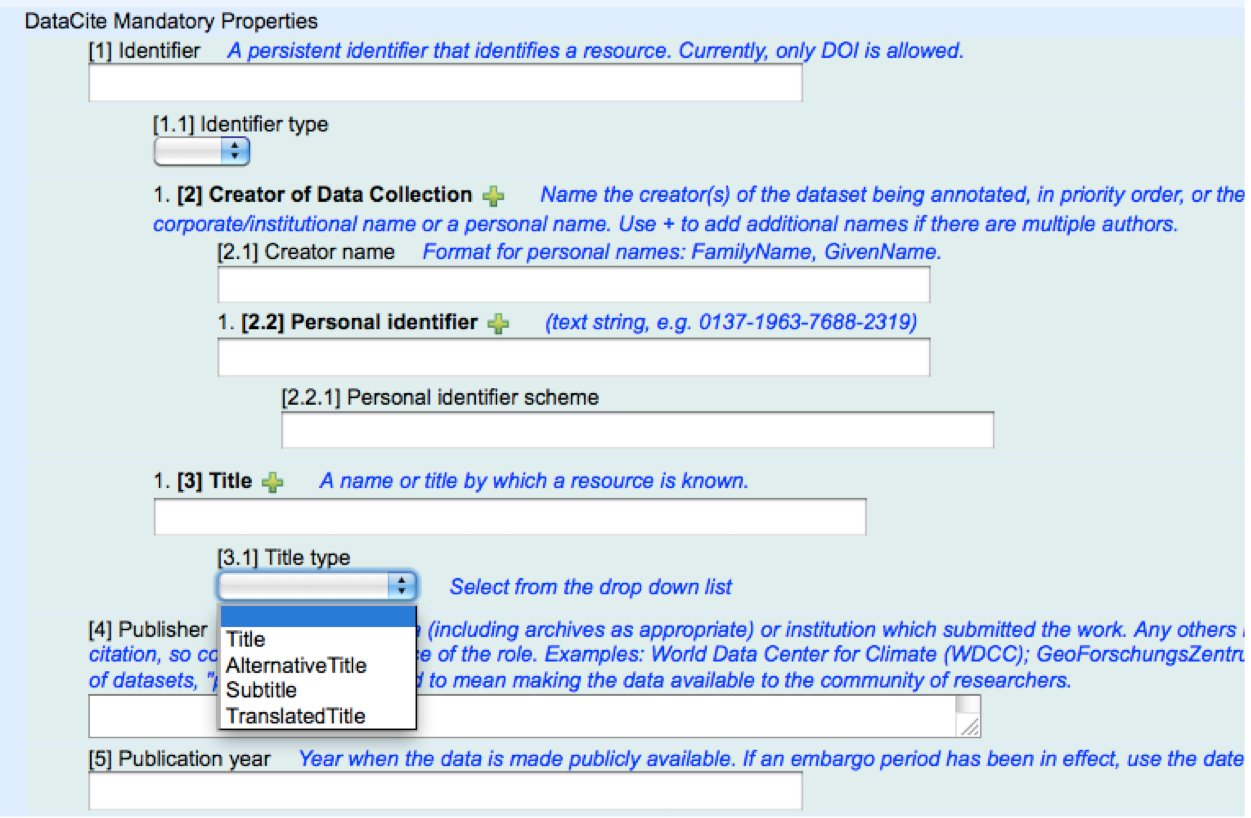
DataCite is an international organization responsible for the DOIs (Digital Object Identifiers) issued for research datasets. For each DOI issued, DataCite requires the data publisher to create and submit to DataCite descriptive metadata that can aid resource discovery.
The purpose of mapping DataCite metadata elements to ontology terms is to enable DataCite metadata to be published in RDF as Open Linked Data, enabling these metadata to be understood programmatically and integrated automatically with similar data from elsewhere.

Benoît Fontaine et al. recently published a study concluding that average lag time between a species being discovered and subsequently described is 21 years. The paper concludes: This is a conclusion that merits more investigation, especially as the title of the paper suggests there is an appalling lack of efficiency (or resources) in the way we decsribe biodiversity.

In a previous blog post, I described the work that Silvio Peroni and I undertook in May 2011 to map the main terms from the DataCite Metadata Kernel v2.0 to RDF. To enable that, we created a ‘proto-ontology’, the DataCite Ontology version 0.2, that contained just the following four object properties: *
The worm that turned (epigenetics) GigaDB, GigaScience ’s associated database, has had a number of new datasets just added, many for data types previously not hosted. Today marks the publication of new research in our sister BMC journal Genome Biology shaking up the epigenetics field by shattering the assumption that DNA methylation is absent in nematodes.
Scientific workflow software such as Taverna, Knime and Pipeline Pilot can overcome interoperability issues relating to the access of tools and data format conversions. Such tasks are automatically handled by the data processing pipeline during its enactment by the workflow software. Galaxy is another workflow system, and its annual conference was held last month at the University of Chicago.