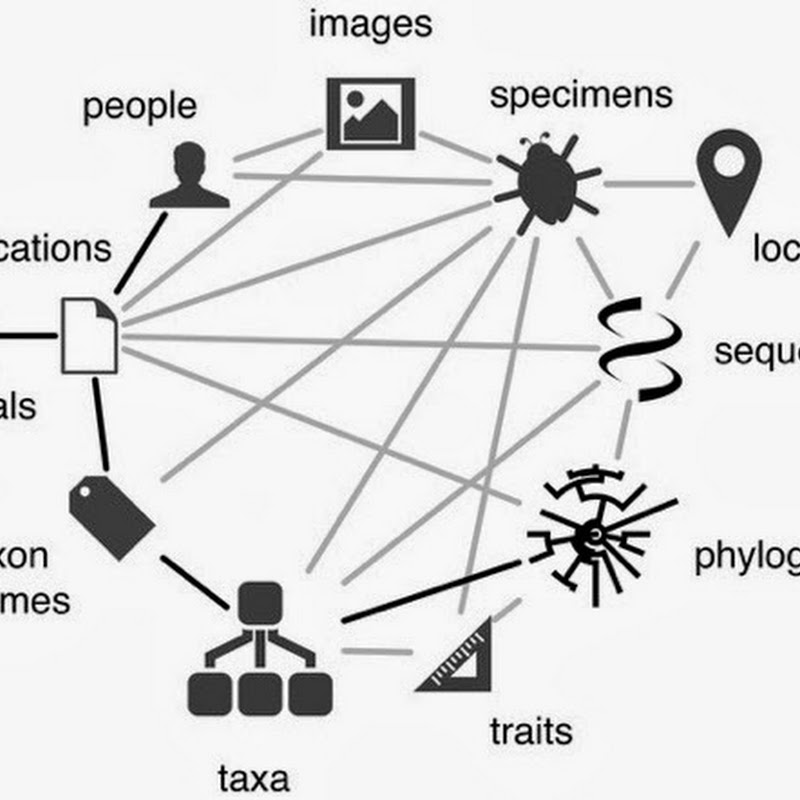
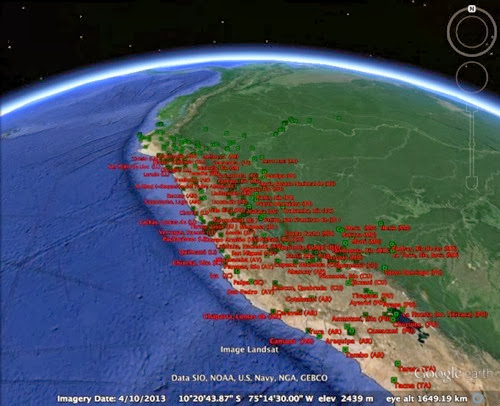
Today I managed to publish some data from a GitHub repository directly to GBIF. Within a few minutes (and with Tim Robertson on hand via Skype to debug a few glitches) the data was automatically indexed by GBIF and its maps updated. You can see the data I uploaded here. The data I uploaded came from this paper: This is the data I used to build the geophylogeny for Banza using Google Earth.