Creating an RSS feed for your Quarto blog can be tricky, but there’s one essential detail that can save you a lot of frustration: matching your file names correctly. The Key to Success: Matching Names When configuring your RSS feed in the _quarto.yml file, ensure that the name you use for the RSS icon matches the name of your listing file. This might seem minor, but it’s crucial for your RSS feed to work properly.
Messaggi di Rogue Scholar
There’s plenty of guides to getting going on Mastodon, aimed at people leaving Twitter. I just wanted to post a couple of technical points about making the switch that might be of interest to people who maintain webpages with Twitter content (feeds, embeds). Mastodon status updates (feed/timeline) Twitter provided a widget that meant that an account’s timeline could be embedded on a website.
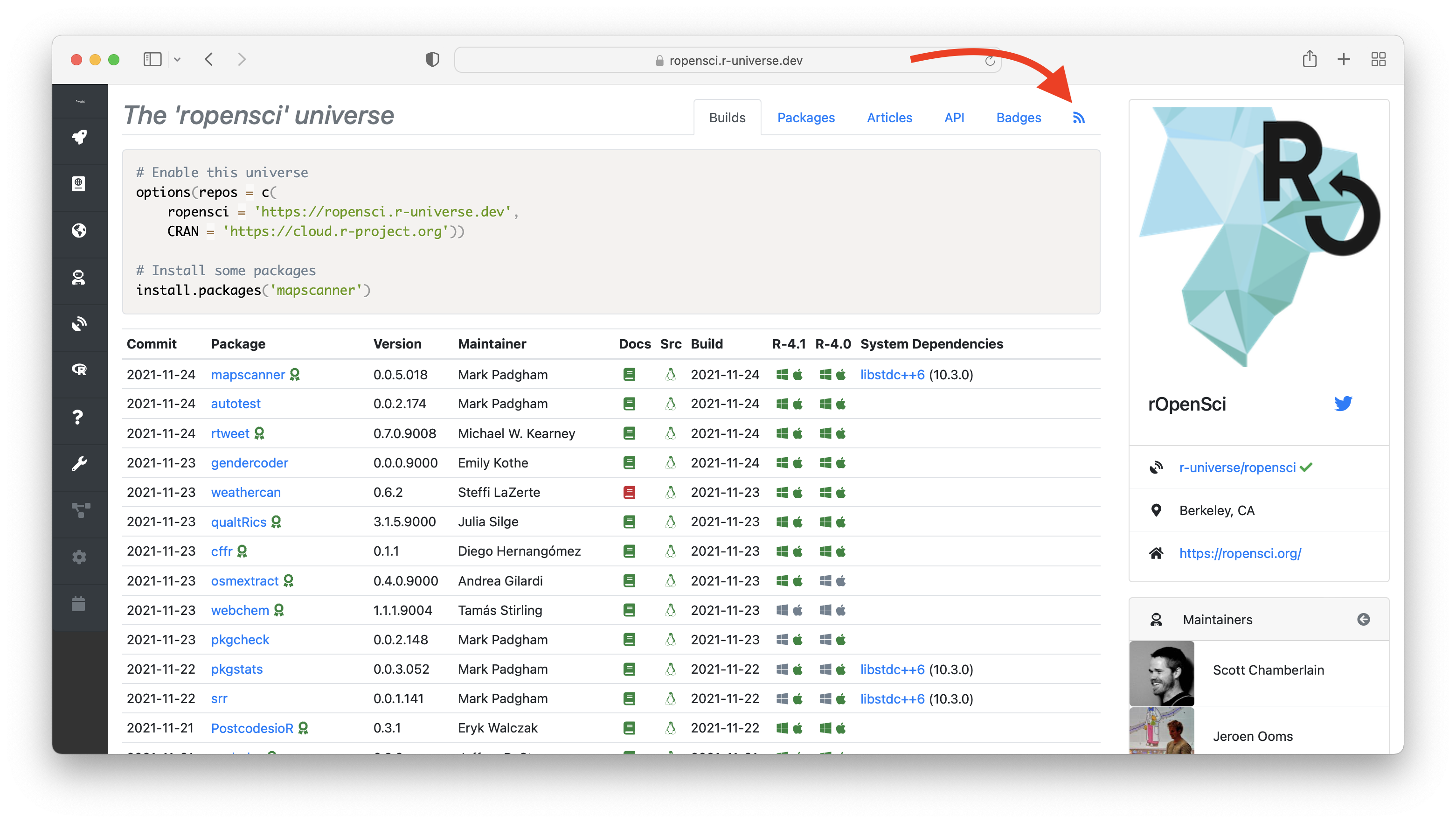
Continuous deployment in r-universe A major difference between r-universe and static repositories like CRAN and BioConductor is continuous deployment: packages in r-universe are continuously built in CI and immediately deployed to our package server. This package server stores binaries and metadata in a database, which enables us to dynamically query and expose all the package data through APIs, dashboards, feeds, etc.
Over a decade ago RSS (RDF Site Summary or Really Simple Syndication) was attracting a lot of interest as a way to integrate data across various websites. Many science publishers would provide a list of their latest articles in XML in one of three flavours of RSS (RDF, RSS, Atom). This led to tools such as uBioRSS [1] and my own e-Biosphere Challenge: visualising biodiversity digitisation in real time.
[Modern scientists are busier than ever. Their typical days are filled not only with experimental work, but also with teaching, supervising, mentoring, grant applications, budget planning… The list goes on and on. No wonder there is barely any time left to stay on top of the field. Keeping track of published literature is made easier by following these simple tips:]{style=“font-family: "arial"; font-size: 11pt;

Following on from (but unrelated to) my post last week about feed tools we have two posts, one from Deepak Singh, and one from Neil Saunders, both talking about ‘friend feeds’ or ‘lifestreams’. The idea here is of aggregating all the content you are generating (or is being generated about you?) into one place. There are a couple of these about but the main ones seem to be Friendfeed and Profiliac.

I can now announce the first closed beta testing phase of an RSS reader intended for scientists. So far, we have something like a Feedly clone with a few extras built in, such as collecting the most tweeted articles of the last 24h, some rudimentary ability to sort/filter either feeds or groups of feeds. It’s not a whole lot, yet, so keep your expectations low We’re just getting started.
If we imagine what the specification for building a scholarly communications system would look like there are some fairly obvious things we would want it to enable. Registration of priority, archival, re-use and replication, and filtering.

Continuing on from my previous post Viewing scientific articles on the iPad: towards a universal article reader, here are some brief notes on the PLoS iPad app that I've previously been critical of. There are two key things to note about this app. The first is that it uses the page turning metaphor. The article is displayed as a PDF, a page at a time, and the user swipes the page to turn it over.

Being in an unusually constructive mood, I've spent the last couple of days playing with the TreeBASE II API, in an effort to find out how hard it would be to replace TreeBASE's frankly ghastly interface. After some hair pulling and bad language I've got something to work. It's very crude, but gives a glimpse at what can be done.